

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140024.11 + phase: 0
(381 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
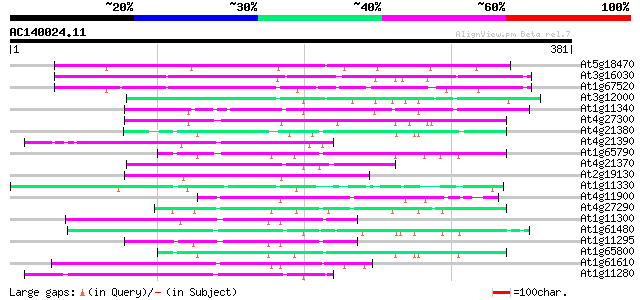
Score E
Sequences producing significant alignments: (bits) Value
At5g18470 unknown protein 135 5e-32
At3g16030 putative receptor kinase 108 6e-24
At1g67520 putative receptor protein kinase 98 7e-21
At3g12000 hypothetical protein 82 6e-16
At1g11340 receptor kinase, putative 80 2e-15
At4g27300 putative receptor protein kinase 73 3e-13
At4g21380 receptor-like serine/threonine protein kinase ARK3 70 3e-12
At4g21390 serine/threonine kinase - like protein 67 1e-11
At1g65790 receptor kinase, putative 67 1e-11
At4g21370 receptor kinase - like protein 66 4e-11
At2g19130 putative receptor-like protein kinase 66 4e-11
At1g11330 receptor-like protein kinase, putative 66 4e-11
At4g11900 KI domain interacting kinase 1 -like protein 65 6e-11
At4g27290 putative receptor like kinase 65 8e-11
At1g11300 putative s-locus protein kinase (PPC:1.7.2) 64 1e-10
At1g61480 S-like receptor protein kinase; member of gene cluster... 62 7e-10
At1g11295 putative s-locus protein kinase (PPC:1.7.2) 58 1e-08
At1g65800 receptor kinase, putative 57 2e-08
At1g61610 56 3e-08
At1g11280 serine/threonine kinase like protein 56 4e-08
>At5g18470 unknown protein
Length = 413
Score = 135 bits (339), Expect = 5e-32
Identities = 104/353 (29%), Positives = 161/353 (45%), Gaps = 44/353 (12%)
Query: 31 SDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSD--------FQCLFISVNADYGKVVWV 82
+D+LKPG +L +L S G F L F +S + L I + +VWV
Sbjct: 30 TDTLKPGQQLRDWEQLISADGIFTLGFFTPKDSSTSELGSAGLRYLGIWPQSIPINLVWV 89
Query: 83 YDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQP----TNNTVATMLDAGNFVL 138
+ S+ ++ LS+D +GVLKI N PI++ P N A +LD GNFV+
Sbjct: 90 GNPTESVSDSSGSLSIDTNGVLKITQANAIPILVNQRPAAQLSLVGNVSAILLDTGNFVV 149
Query: 139 QQFLPNGSMS-VLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVS---------DKFNLEW 188
++ P G VLWQSFD+P++ L+P MK+G N +T S+ S F L
Sbjct: 150 REIRPGGVPGRVLWQSFDHPTNTLLPGMKIGFNLRTKKEVSVTSWITDQVPVPGAFRLGL 209
Query: 189 EPK-QGELNIKKSGKVYWKSGKLKSNGLFENIPANVQSR---YQYIIVSNKDEDSFTFEV 244
+P +L + + G++YW SG L +NG ++ V Y++ SNK F++ +
Sbjct: 210 DPSGANQLLVWRRGEIYWSSGILTNNG-SSHLNLEVSRHYIDYEFKFDSNKYMKYFSYSI 268
Query: 245 KDGK---FAQWELSSKGKLVGDDGYIANADMCYGYNSDGGCQK----------WEDIPTC 291
K F+ W L + G++ +N + S C+ E C
Sbjct: 269 KKANSSVFSSWFLDTLGQITVTFSLSSNNSSTWISESSEPCKTDLKNSSAICITEKPTAC 328
Query: 292 REPGEMFQKKAGRPSIDNSTTYEF----DVTYSYSDCKIRCWKNCSCNGFQLY 340
R+ E F+ + G +N+ Y F ++ SDC CW+NCSC FQ +
Sbjct: 329 RKGSEYFEPRRGYMMENNTGYYPFYYDDSLSAGLSDCHGTCWRNCSCIAFQAF 381
>At3g16030 putative receptor kinase
Length = 850
Score = 108 bits (269), Expect = 6e-24
Identities = 106/380 (27%), Positives = 158/380 (40%), Gaps = 64/380 (16%)
Query: 31 SDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNADY-GKVVWVYDINHSI 89
+D+L G L +L S F L+F N NS L I N Y VW+ + N+ +
Sbjct: 24 TDTLLQGQYLKDGQELVSAFNIFKLKFFNFENSSNWYLGIWYNNFYLSGAVWIANRNNPV 83
Query: 90 DFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLPNGSMS- 148
+ L++D G L+I + ++ SS + T NT +LD+GN LQ+ +GSM
Sbjct: 84 LGRSGSLTVDSLGRLRI-LRGASSLLELSSTETTGNTTLKLLDSGNLQLQEMDSDGSMKR 142
Query: 149 VLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLV-----------SDKFNLEWEPKQGELNI 197
LWQSFDYP+D L+P MKLG N KTG W L S F ++ + L I
Sbjct: 143 TLWQSFDYPTDTLLPGMKLGFNVKTGKRWELTSWLGDTLPASGSFVFGMD-DNITNRLTI 201
Query: 198 KKSGKVYWKSGKLKSNGL-FENIPANVQSRYQYIIVSNKDEDSFTFEVKDG----KFAQW 252
G VYW SG G E + N + + VS + E F + + F +
Sbjct: 202 LWLGNVYWASGLWFKGGFSLEKLNTN---GFIFSFVSTESEHYFMYSGDENYGGPLFPRI 258
Query: 253 ELSSKGKL--VGDDG---YIANADMCYGYNSDGGC------------------------- 282
+ +G L + DG ++ + +G + GC
Sbjct: 259 RIDQQGSLQKINLDGVKKHVHCSPSVFGEELEYGCYQQNFRNCVPARYKEVTGSWDCSPF 318
Query: 283 -------QKWEDIPTCREPGEMFQKKAGRPSIDNSTTY-EFDVTYSYSDCKIRCWKNCSC 334
+K D+ C G F++ PS +N + E S DC ++C +NCSC
Sbjct: 319 GFGYTYTRKTYDLSYCSRFGYTFRETVS-PSAENGFVFNEIGRRLSSYDCYVKCLQNCSC 377
Query: 335 NGFQLYYSNMTGCVFLSWNS 354
+ + TGC WN+
Sbjct: 378 VAYASTNGDGTGCEI--WNT 395
>At1g67520 putative receptor protein kinase
Length = 833
Score = 98.2 bits (243), Expect = 7e-21
Identities = 99/351 (28%), Positives = 152/351 (43%), Gaps = 43/351 (12%)
Query: 31 SDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSD-------FQCLFISVNADYGKVVWVY 83
+D+L G L +L S F L+F N NS+ F L+++ ++ + VW+
Sbjct: 24 TDTLHQGQFLKDGQELVSAFKIFKLKFFNFKNSENLYLGIWFNNLYLNTDSQ-DRPVWIA 82
Query: 84 DINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLP 143
+ N+ I + L++D G LKI + ++ SS + T NT +LD+GN LQ+
Sbjct: 83 NRNNPISDRSGSLTVDSLGRLKI-LRGASTMLELSSIETTRNTTLQLLDSGNLQLQEMDA 141
Query: 144 NGSMS-VLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGE-------- 194
+GSM VLWQSFDYP+D L+P MKLG + KT W L S + + P G
Sbjct: 142 DGSMKRVLWQSFDYPTDTLLPGMKLGFDGKTRKRWELTS--WLGDTLPASGSFVFGMDTN 199
Query: 195 ----LNIKKSGKVYWKSGKLKSNGLFENIPANVQSRYQYIIVSNKDEDSFTFEVKDGKFA 250
L I G +YW SG L + G F N + + + VS K F + G
Sbjct: 200 ITNVLTILWRGNMYWSSG-LWNKGRFSEEELN-ECGFLFSFVSTKSGQYFMY---SGDQD 254
Query: 251 QWELSSKGKLVGDDGYIANADMCYGYNSDGGCQKWEDIPTCREPG-----EMFQKKAGRP 305
++ + G + M N + C G E + + R
Sbjct: 255 DARTFFPTIMIDEQGILRREQMHRQRNRQNYRNR-----NCLAAGYVVRDEPYGFTSFRV 309
Query: 306 SIDNSTTYEFDV--TYSYSDCKIRCWKNCSCNGFQLYYSNMTGCVFLSWNS 354
++ +S + F + T+S DC C +N SC + + TGC WN+
Sbjct: 310 TVSSSASNGFVLSGTFSSVDCSAICLQNSSCLAYASTEPDGTGCEI--WNT 358
>At3g12000 hypothetical protein
Length = 439
Score = 81.6 bits (200), Expect = 6e-16
Identities = 79/344 (22%), Positives = 140/344 (39%), Gaps = 69/344 (20%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQ 139
VWV + ++ + + L + Y+ ++ ++ + + VA +LD GNFVL+
Sbjct: 88 VWVANRDNPLSKSIGTLKISYANLVLLDHSGTLVWSTNLTRTVKSPVVAELLDNGNFVLR 147
Query: 140 QFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNIK- 198
N LWQSFDYP D L+P MK+G + KTGH L S + ++P G+ + K
Sbjct: 148 DSKGNYQNRFLWQSFDYPVDTLLPEMKIGRDLKTGHETFLSS--WRSPYDPSSGDFSFKL 205
Query: 199 -----------KSGKVYWKSGKLKSNGLFENIPANVQSRYQYII---VSNKDEDSFTFEV 244
K + ++SG G F IP Y ++ + N+ E +++F+V
Sbjct: 206 GTQGLPEFYLFKKEFLLYRSGPWNGVG-FSGIPTMQNWSYFDVVNNFIENRGEVAYSFKV 264
Query: 245 KDG---------------KFAQWELSSKG---------------KLVGDDGYI-----AN 269
D + ++W+ +S ++ G D Y
Sbjct: 265 TDHSMHYVRFTLTTERLLQISRWDTTSSEWNLFGVLPTEKCDLYQICGRDSYCDTKTSPT 324
Query: 270 ADMCYGY-----------NSDGGCQKWEDIPTCREPGEMFQKKAGRPSIDNSTTYEFDVT 318
+ G+ ++ GC + + C G K R + ++T D T
Sbjct: 325 CNCIKGFVPKNVTAWALGDTFEGCVRKSRL-NCHRDGFFLLMK--RMKLPGTSTAIVDKT 381
Query: 319 YSYSDCKIRCWKNCSCNGF--QLYYSNMTGCVFLSWNSTQYVDM 360
++CK RC K+C+C GF + + +GCV + + Y ++
Sbjct: 382 IGLNECKERCSKDCNCTGFANKDIQNGGSGCVIWTGGAHGYEEL 425
>At1g11340 receptor kinase, putative
Length = 901
Score = 80.1 bits (196), Expect = 2e-15
Identities = 83/343 (24%), Positives = 139/343 (40%), Gaps = 78/343 (22%)
Query: 79 VVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSP------QPTNNTVATMLD 132
+VWV + +H I+ + ++ G L + + + + +I+S+ +PT VAT+ D
Sbjct: 136 IVWVANRDHPINDTSGMVKFSNRGNLSVYASDNETELIWSTNVSDSMLEPT--LVATLSD 193
Query: 133 AGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQ 192
GN VL F P S W+SFD+P+D +P M+LG RK G + SL S K + +P
Sbjct: 194 LGNLVL--FDPVTGRS-FWESFDHPTDTFLPFMRLGFTRKDGLDRSLTSWKSH--GDPGS 248
Query: 193 GELNIK------------KSGKVYWKSGKLKSNGLFENIPANVQSRYQYIIVSNKDEDSF 240
G+L ++ K +W+ G + + + V+N+DE SF
Sbjct: 249 GDLILRMERRGFPQLILYKGVTPWWRMGSWTGHRWSGVPEMPIGYIFNNSFVNNEDEVSF 308
Query: 241 TFEVKD------------GKFAQWELSSKGKLVGDDGYIANADMCYGY------------ 276
T+ V D G ++ ++ K +D + + C Y
Sbjct: 309 TYGVTDASVITRTMVNETGTMHRFTWIARDKR-WNDFWSVPKEQCDNYAHCGPNGYCDSP 367
Query: 277 ------------------------NSDGGCQKWEDIPTCREPGEMFQKKAGRPSIDNSTT 312
+S GGC K + C E + K R I +++
Sbjct: 368 SSKTFECTCLPGFEPKFPRHWFLRDSSGGCTKKKRASICSEKDGFVKLK--RMKIPDTSD 425
Query: 313 YEFDVTYSYSDCKIRCWKNCSCNGF-QLYYSNMTGCV-FLSWN 353
D+ + +CK RC KNCSC + Y+ + G + L W+
Sbjct: 426 ASVDMNITLKECKQRCLKNCSCVAYASAYHESKRGAIGCLKWH 468
>At4g27300 putative receptor protein kinase
Length = 815
Score = 72.8 bits (177), Expect = 3e-13
Identities = 84/321 (26%), Positives = 131/321 (40%), Gaps = 66/321 (20%)
Query: 79 VVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSP-----QPTNNTVATMLDA 133
VVWV + N+ + + L+L G L++ K + SS + NN + + +
Sbjct: 75 VVWVANRNNPLYGTSGFLNLSSLGDLQLFDGEHKALWSSSSSSTKASKTANNPLLKISCS 134
Query: 134 GNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDK---------F 184
GN + +G +VLWQSFDYP + ++ MKLG N KT WSL S K F
Sbjct: 135 GNLISS----DGEEAVLWQSFDYPMNTILAGMKLGKNFKTQMEWSLSSWKTLKDPSPGDF 190
Query: 185 NLEWEPK-QGELNIKKSGKVYWKSGKLKSNGL-FENIPA--NVQSRYQYIIVSNKDEDSF 240
L + + +L ++K+G + NGL F PA S + Y S+ E ++
Sbjct: 191 TLSLDTRGLPQLILRKNGDSSYSYRLGSWNGLSFTGAPAMGRENSLFDYKFTSSAQEVNY 250
Query: 241 TFEVKDGKFAQWELSSKGKL------VGDDGYIANA---DMCYGYNSDGGC--------- 282
++ + ++ L++ GKL + +AN D C Y+ G
Sbjct: 251 SWTPRHRIVSRLVLNNTGKLHRFIQSKQNQWILANTAPEDECDYYSICGAYAVCGINSKN 310
Query: 283 ---------------QKW----------EDIPTCREPGEMFQKKAGRPSIDNS-TTYEFD 316
+KW +IPT E + F K G D S + Y+
Sbjct: 311 TPSCSCLQGFKPKSGRKWNISRGAYGCVHEIPTNCEKKDAFVKFPGLKLPDTSWSWYDAK 370
Query: 317 VTYSYSDCKIRCWKNCSCNGF 337
+ DCKI+C NCSC +
Sbjct: 371 NEMTLEDCKIKCSSNCSCTAY 391
>At4g21380 receptor-like serine/threonine protein kinase ARK3
Length = 850
Score = 69.7 bits (169), Expect = 3e-12
Identities = 80/334 (23%), Positives = 132/334 (38%), Gaps = 93/334 (27%)
Query: 78 KVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNT----------- 126
K +V+ N ++S+ G LKI N +++ S P +T
Sbjct: 77 KRTYVWVANRDTPLSSSI------GTLKISDSNL--VVLDQSDTPVWSTNLTGGDVRSPL 128
Query: 127 VATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNL 186
VA +LD GNFVL+ + VLWQSFD+P+D L+P MKLG + KTG N +F
Sbjct: 129 VAELLDNGNFVLRDSKNSAPDGVLWQSFDFPTDTLLPEMKLGWDAKTGFN------RFIR 182
Query: 187 EW----EPKQGELNIKKSGKVY------------WKSGKLKSNGL-FENIPANVQSRYQ- 228
W +P G+ + K + + ++SG NG+ F +P Y
Sbjct: 183 SWKSPDDPSSGDFSFKLETEGFPEIFLWNRESRMYRSGPW--NGIRFSGVPEMQPFEYMV 240
Query: 229 YIIVSNKDEDSFTFEV-KDGKFAQWELSSKGKLV----------GDDGYIANADMC---- 273
+ ++K+E +++F + K +++ +SS G L + + A D C
Sbjct: 241 FNFTTSKEEVTYSFRITKSDVYSRLSISSSGLLQRFTWIETAQNWNQFWYAPKDQCDEYK 300
Query: 274 ----YGY--------------------------NSDGGCQKWEDIPTCREPGEMFQKKAG 303
YGY + GC + + G + KK
Sbjct: 301 ECGVYGYCDSNTSPVCNCIKGFKPRNPQVWGLRDGSDGCVRKTLLSCGGGDGFVRLKKMK 360
Query: 304 RPSIDNSTTYEFDVTYSYSDCKIRCWKNCSCNGF 337
P ++TT D +C+ +C ++C+C F
Sbjct: 361 LP---DTTTASVDRGIGVKECEQKCLRDCNCTAF 391
>At4g21390 serine/threonine kinase - like protein
Length = 849
Score = 67.4 bits (163), Expect = 1e-11
Identities = 63/224 (28%), Positives = 105/224 (46%), Gaps = 21/224 (9%)
Query: 11 VLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFI 70
+ +Y +L+ ++ A + +SL+ G +N+K L S Q F L F + +S + L I
Sbjct: 13 LFLYFFLYESSMAANTIRR-GESLRDG--INHKP-LVSPQKTFELGFFSPGSSTHRFLGI 68
Query: 71 SV-NADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPII---IYSSPQPTNNT 126
N + VVWV + I + VL + G L + + I SS NN
Sbjct: 69 WYGNIEDKAVVWVANRATPISDQSGVLMISNDGNLVLLDGKNITVWSSNIESSTTNNNNR 128
Query: 127 VATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNL 186
V ++ D GNFVL + + +W+SF++P+D +P M++ VN +TG N + VS +
Sbjct: 129 VVSIHDTGNFVLSE---TDTDRPIWESFNHPTDTFLPQMRVRVNPQTGDNHAFVSWRSET 185
Query: 187 EWEPKQGELNIKKSGK---VYWKSGKLK-------SNGLFENIP 220
+ P L + SG V W+ K + ++ +F IP
Sbjct: 186 DPSPGNYSLGVDPSGAPEIVLWEGNKTRKWRSGQWNSAIFTGIP 229
Score = 32.0 bits (71), Expect = 0.58
Identities = 14/31 (45%), Positives = 18/31 (57%), Gaps = 4/31 (12%)
Query: 323 DCKIRCWKNCSCNGFQLYYSNMTGCVFLSWN 353
DC+ RC +NCSCN YS + G + WN
Sbjct: 380 DCRERCLRNCSCNA----YSLVGGIGCMIWN 406
>At1g65790 receptor kinase, putative
Length = 843
Score = 67.4 bits (163), Expect = 1e-11
Identities = 74/302 (24%), Positives = 123/302 (40%), Gaps = 73/302 (24%)
Query: 101 SGVLKIESQNRKPIIIYSSPQPTNNT-----------VATMLDAGNFVLQQFLPNGSMSV 149
+G LKI N +I S +P +T A +LD GNF+L+ + + +
Sbjct: 92 NGTLKISGNNL--VIFDQSDRPVWSTNITGGDVRSPVAAELLDNGNFLLR----DSNNRL 145
Query: 150 LWQSFDYPSDVLIPMMKLGVNRKTGHN-----WSLVSDKFNLEWEPKQ-----GELNIKK 199
LWQSFD+P+D L+ MKLG ++KTG N W D + E+ K E I
Sbjct: 146 LWQSFDFPTDTLLAEMKLGWDQKTGFNRILRSWKTTDDPSSGEFSTKLETSEFPEFYICS 205
Query: 200 SGKVYWKSGKLKSNGL-FENIPANVQSRYQ-YIIVSNKDEDSFTFEV-KDGKFAQWELSS 256
+ ++SG NG+ F ++P +Q Y Y ++K+E ++++ + K +++ L+S
Sbjct: 206 KESILYRSGPW--NGMRFSSVPGTIQVDYMVYNFTASKEEVTYSYRINKTNLYSRLYLNS 263
Query: 257 KGKL----------VGDDGYIANADMCYGYNSDG--GCQKWEDIPTC----------REP 294
G L + + D+C Y G G +P C +
Sbjct: 264 AGLLQRLTWFETTQSWKQLWYSPKDLCDNYKVCGNFGYCDSNSLPNCYCIKGFKPVNEQA 323
Query: 295 GEMFQKKAG-------------------RPSIDNSTTYEFDVTYSYSDCKIRCWKNCSCN 335
++ AG R + ++T D CK RC ++C+C
Sbjct: 324 WDLRDGSAGCMRKTRLSCDGRDGFTRLKRMKLPDTTATIVDREIGLKVCKERCLEDCNCT 383
Query: 336 GF 337
F
Sbjct: 384 AF 385
>At4g21370 receptor kinase - like protein
Length = 844
Score = 65.9 bits (159), Expect = 4e-11
Identities = 53/199 (26%), Positives = 94/199 (46%), Gaps = 21/199 (10%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQ 139
VWV + + + +L + + ++ +++ + + ++ VA +LD GNFVL+
Sbjct: 83 VWVANRDTPLSNPIGILKISNANLVILDNSDTHVWSTNLTGAVRSSVVAELLDNGNFVLR 142
Query: 140 QFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNIK- 198
N S LWQSFD+P+D L+P MKLG + K G N + S K + ++P G K
Sbjct: 143 GSKINESDEFLWQSFDFPTDTLLPQMKLGRDHKRGLNRFVTSWKSS--FDPSSGSFMFKL 200
Query: 199 -----------KSGKVYWKSGK---LKSNGLFENIPANVQSRYQYIIVSNKDEDSFTFEV 244
S ++SG L+ +G+ E Y N++E ++TF V
Sbjct: 201 ETLGLPEFFGFTSFLEVYRSGPWDGLRFSGILE---MQQWDDIIYNFTENREEVAYTFRV 257
Query: 245 KD-GKFAQWELSSKGKLVG 262
D +++ +++ G+L G
Sbjct: 258 TDHNSYSRLTINTVGRLEG 276
>At2g19130 putative receptor-like protein kinase
Length = 828
Score = 65.9 bits (159), Expect = 4e-11
Identities = 45/180 (25%), Positives = 87/180 (48%), Gaps = 14/180 (7%)
Query: 79 VVWVYDINHSI-DFNTSVLSLDYSGVLKIESQNRKPIII--YSSPQPTNNTVATMLDAGN 135
++WV + + ++ D N+SV + ++ ++ + P+ +S + A + D GN
Sbjct: 71 ILWVANRDKAVSDKNSSVFKISNGNLILLDGNYQTPVWSTGLNSTSSVSALEAVLQDDGN 130
Query: 136 FVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDK---------FNL 186
VL+ + S +VLWQSFD+P D +P +K+ ++++TG + L S K F+L
Sbjct: 131 LVLRTGGSSLSANVLWQSFDHPGDTWLPGVKIRLDKRTGKSQRLTSWKSLEDPSPGLFSL 190
Query: 187 EWEPKQGELNIKKSGKVYWKSGKLK-SNGLFENIP-ANVQSRYQYIIVSNKDEDSFTFEV 244
E + + YW SG + +F+++P + Y + SN + FT+ +
Sbjct: 191 ELDESTAYKILWNGSNEYWSSGPWNPQSRIFDSVPEMRLNYIYNFSFFSNTTDSYFTYSI 250
>At1g11330 receptor-like protein kinase, putative
Length = 840
Score = 65.9 bits (159), Expect = 4e-11
Identities = 88/357 (24%), Positives = 144/357 (39%), Gaps = 69/357 (19%)
Query: 1 MISFEIKKQVVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNN 60
++S I+++ VL+ L + +C + K + L K G F F
Sbjct: 2 VVSVTIRRRFVLLLLACTCLLSRRLCFGEDRITFSSPIKDSESETLLCKSGIFRFGFFTP 61
Query: 61 SNSDFQCLFISV---NADYGKVVWVYDINHSIDFNTSVLSLDYSGVLKI-ESQNRKPIII 116
NS + ++ + VVWV + + I+ + V+S+ G L + + +NR ++
Sbjct: 62 VNSTTRLRYVGIWYEKIPIQTVVWVANKDSPINDTSGVISIYQDGNLAVTDGRNR---LV 118
Query: 117 YSS----PQPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRK 172
+S+ P N T ++D+GN +LQ NG +LW+SF +P D +P M LG + +
Sbjct: 119 WSTNVSVPVAPNATWVQLMDSGNLMLQDNRNNGE--ILWESFKHPYDSFMPRMTLGTDGR 176
Query: 173 TGHNWSLVSDKFNLEWEPKQG------------ELNIKKSGKVYWKSGKLKSNGLFENIP 220
TG N L S + +P G EL I K+ W+SG P
Sbjct: 177 TGGNLKLTS--WTSHDDPSTGNYTAGIAPFTFPELLIWKNNVPTWRSG-----------P 223
Query: 221 ANVQSRYQYIIVSNKDEDSFTFEVKDGKFAQWELSSKGKLVGDDGYIANADMCYGYNSDG 280
N Q + + + DS F DG + L+S + Y AN Y +N D
Sbjct: 224 WNGQ-----VFIGLPNMDSLLF--LDG----FNLNSDNQGTISMSY-ANDSFMYHFNLD- 270
Query: 281 GCQKWEDIPTCREPGEMFQKKAGRPSIDNSTTYEFDVTYSYSDCKI--RCWKNCSCN 335
G ++QK + T+ V + Y+DC RC + SC+
Sbjct: 271 ------------PEGIIYQKDWS----TSMRTWRIGVKFPYTDCDAYGRCGRFGSCH 311
>At4g11900 KI domain interacting kinase 1 -like protein
Length = 849
Score = 65.1 bits (157), Expect = 6e-11
Identities = 62/225 (27%), Positives = 100/225 (43%), Gaps = 33/225 (14%)
Query: 128 ATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSD----K 183
A + D+GN VL+ PN S +VLWQSFD+PSD +P K+ + + +W + D +
Sbjct: 157 AVLFDSGNLVLRDG-PNSSAAVLWQSFDHPSDTWLPGGKIRLGSQLFTSWESLIDPSPGR 215
Query: 184 FNLEWEPKQGEL-NIKKSGKVYWKSGKLKS-NGLFENIPANVQSRYQYIIVSNKDEDSFT 241
++LE++PK L + K YW SG L F+ P ++ + + N DE T
Sbjct: 216 YSLEFDPKLHSLVTVWNRSKSYWSSGPLYDWLQSFKGFPELQGTKLSFTL--NMDESYIT 273
Query: 242 FEVKDGKFAQWELSSKGKLVGDDGYI----------ANADMCYGYNSDGG---CQKWEDI 288
F V + + G+ + ++ + C YNS G C + +
Sbjct: 274 FSVDPQSRYRLVMGVSGQFMLQVWHVDLQSWRVILSQPDNRCDVYNSCGSFGICNENREP 333
Query: 289 PTCR-EPGEMFQKKAGRPSIDNSTTYEFDVTYSYSDCKIRCWKNC 332
P CR PG F+++ + S D+S Y CK + +C
Sbjct: 334 PPCRCVPG--FKREFSQGS-DDSNDYS-------GGCKRETYLHC 368
>At4g27290 putative receptor like kinase
Length = 772
Score = 64.7 bits (156), Expect = 8e-11
Identities = 75/310 (24%), Positives = 122/310 (39%), Gaps = 78/310 (25%)
Query: 99 DYSGVLKIESQ-------NRKPIIIYSSPQPTN------NTVATMLDAGNFVLQQFLPNG 145
D SG LK+ +R II SS P++ N + +LD GN V++
Sbjct: 74 DLSGTLKVSENGSLCLFNDRNHIIWSSSSSPSSQKASLRNPIVQILDTGNLVVRN--SGD 131
Query: 146 SMSVLWQSFDYPSDVLIPMMKLGVNRKTGHN-----WSLVSDKFNLEWEPKQG-----EL 195
+WQS DYP D+ +P MK G+N TG N W + D + K +
Sbjct: 132 DQDYIWQSLDYPGDMFLPGMKYGLNFVTGLNRFLTSWRAIDDPSTGNYTNKMDPNGVPQF 191
Query: 196 NIKKSGKVYWKSGKLKSNGL----FENIPANVQSRYQYIIVSNKDEDSFTFEVKD-GKFA 250
+KK+ V +++G NGL N+ N RY+Y+ ++E +T+++++
Sbjct: 192 FLKKNSVVVFRTGPW--NGLRFTGMPNLKPNPIYRYEYVF--TEEEVYYTYKLENPSVLT 247
Query: 251 QWELSSKG---------KLVGDDGYI-ANADMCYGY---NSDGGCQKWEDIPTCR----- 292
+ +L+ G L + Y+ A D C Y S G C E P CR
Sbjct: 248 RMQLNPNGALQRYTWVDNLQSWNFYLSAMMDSCDQYTLCGSYGSCNINES-PACRCLKGF 306
Query: 293 -------------------------EPGEMFQKKAGRPSIDNSTTYEFDVTYSYSDCKIR 327
GE K + + ++ T +D ++CK
Sbjct: 307 VAKTPQAWVAGDWSEGCVRRVKLDCGKGEDGFLKISKLKLPDTRTSWYDKNMDLNECKKV 366
Query: 328 CWKNCSCNGF 337
C +NC+C+ +
Sbjct: 367 CLRNCTCSAY 376
>At1g11300 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 63.9 bits (154), Expect = 1e-10
Identities = 57/215 (26%), Positives = 93/215 (42%), Gaps = 21/215 (9%)
Query: 39 KLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNA-DYGKVVWVYDINHSIDFNTSVLS 97
KLN + S F F + NS + I N+ V+WV + + I+ ++ V+S
Sbjct: 35 KLNDSETIVSSFRTFRFGFFSPVNSTSRYAGIWYNSVSVQTVIWVANKDKPINDSSGVIS 94
Query: 98 LDYSGVLKIESQNRKPI--IIYSSPQPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFD 155
+ G L + R+ + S+ N+TVA +LD+GN VL++ S + LW+SF
Sbjct: 95 VSQDGNLVVTDGQRRVLWSTNVSTQASANSTVAELLDSGNLVLKEA---SSDAYLWESFK 151
Query: 156 YPSDVLIPMMKLGVNRKTG------HNWSLVSD--------KFNLEWEPKQGELNIKKSG 201
YP+D +P M +G N + G +W SD L P+ +N +
Sbjct: 152 YPTDSWLPNMLVGTNARIGGGNVTITSWKSPSDPSPGSYTAALVLAAYPELFIMNNNNNN 211
Query: 202 KVYWKSGKLKSNGLFENIPANVQSRYQYIIVSNKD 236
W+SG +F +P + Y + N D
Sbjct: 212 STVWRSGPWNGQ-MFNGLPDVYAGVFLYRFIVNDD 245
>At1g61480 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 809
Score = 61.6 bits (148), Expect = 7e-10
Identities = 88/373 (23%), Positives = 140/373 (36%), Gaps = 70/373 (18%)
Query: 40 LNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNADYGKVV-WVYDINHSIDFNTSVLSL 98
L+ L S G + L F + +NS Q + I +VV WV + + + + L++
Sbjct: 32 LSIGKTLSSSNGVYELGFFSFNNSQNQYVGIWFKGIIPRVVVWVANREKPVTDSAANLTI 91
Query: 99 DYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPS 158
+G L + ++N + +N + A + D GN V+ + N S LW+SF++
Sbjct: 92 SSNGSLLLFNENHSVVWSIGETFASNGSRAELTDNGNLVV---IDNNSGRTLWESFEHFG 148
Query: 159 DVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNIK------------KSGKVYWK 206
D ++P L N TG L S K + +P G+ ++ + K YW+
Sbjct: 149 DTMLPFSNLMYNLATGEKRVLTSWKSHT--DPSPGDFTVQITPQVPSQACTMRGSKTYWR 206
Query: 207 SGKLKSNGLFENIPANVQSRYQYIIVSNKDED---SFTFEVKDGKFAQWELSSKGKLV-- 261
SG + F IP + Y +D + SFT+ ++ K + ++S+G L
Sbjct: 207 SGPW-AKTRFTGIPV-MDDTYTSPFSLQQDTNGSGSFTYFERNFKLSYIMITSEGSLKIF 264
Query: 262 ---GDD---GYIANADMC--YGYNSDGGCQKWEDIPTCR-----EPGEMFQKKAGR---- 304
G D + A + C YG+ G P C+ P + + K G
Sbjct: 265 QHNGMDWELNFEAPENSCDIYGFCGPFGICVMSVPPKCKCFKGFVPKSIEEWKRGNWTDG 324
Query: 305 ------------------------PSIDNSTTYEFDVTYSYSDCKIRCWKNCSCNGFQLY 340
+I YEF C C NCSC F
Sbjct: 325 CVRHTELHCQGNTNGKTVNGFYHVANIKPPDFYEFASFVDAEGCYQICLHNCSCLAFA-- 382
Query: 341 YSNMTGCVFLSWN 353
Y N GC L WN
Sbjct: 383 YINGIGC--LMWN 393
>At1g11295 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 57.8 bits (138), Expect = 1e-08
Identities = 50/177 (28%), Positives = 83/177 (46%), Gaps = 26/177 (14%)
Query: 79 VVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPT----NNTVATMLDAG 134
V+WV + + I+ ++ V+S+ G L + R+ +++S+ T N+TVA +L++G
Sbjct: 76 VIWVANKDTPINDSSGVISISEDGNLVVTDGQRR--VLWSTNVSTRASANSTVAELLESG 133
Query: 135 NFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTG------HNWSLVSD------ 182
N VL+ + + LW+SF YP+D +P M +G N +TG +W+ SD
Sbjct: 134 NLVLKDA---NTDAYLWESFKYPTDSWLPNMLVGTNARTGGGNITITSWTNPSDPSPGSY 190
Query: 183 --KFNLEWEPKQGELNIKKSGKVYWKSGKLKSNGL-FENIPANVQSRYQYIIVSNKD 236
L P+ N + W+SG NGL F +P + Y N D
Sbjct: 191 TAALVLAPYPELFIFNNNDNNATVWRSGPW--NGLMFNGLPDVYPGLFLYRFKVNDD 245
>At1g65800 receptor kinase, putative
Length = 847
Score = 56.6 bits (135), Expect = 2e-08
Identities = 69/301 (22%), Positives = 109/301 (35%), Gaps = 67/301 (22%)
Query: 101 SGVLKIESQNRKPIIIYSSPQPTNNT-----------VATMLDAGNFVLQQFLPNGSMSV 149
+G LKI N +I S +P +T A +LD GNFVL+ N
Sbjct: 92 NGTLKISDNNL--VIFDQSDRPVWSTNITGGDVRSPVAAELLDYGNFVLRDSKNNKPSGF 149
Query: 150 LWQSFDYPSDVLIPMMKLGVNRKTG------HNWSLVSDKFNLEWEPKQ-----GELNIK 198
LWQSFD+P+D L+ MK+G + K+G +W D + ++ K E I
Sbjct: 150 LWQSFDFPTDTLLSDMKMGWDNKSGGFNRILRSWKTTDDPSSGDFSTKLRTSGFPEFYIY 209
Query: 199 KSGKVYWKSGKLKSNGLFENIPANVQSRY-QYIIVSNKDEDSFTFEV-KDGKFAQWELSS 256
+ ++SG N F ++P Y N + +++ V K ++ LSS
Sbjct: 210 NKESITYRSGPWLGN-RFSSVPGMKPVDYIDNSFTENNQQVVYSYRVNKTNIYSILSLSS 268
Query: 257 KGKL----------VGDDGYIANADMC--------YGY---NSDGGCQKWEDIPTCREPG 295
G L + + D+C YGY N+ C + E
Sbjct: 269 TGLLQRLTWMEAAQSWKQLWYSPKDLCDNYKECGNYGYCDANTSPICNCIKGFEPMNEQA 328
Query: 296 EMFQKKAG-------------------RPSIDNSTTYEFDVTYSYSDCKIRCWKNCSCNG 336
+ G + + ++T D +C+ RC K C+C
Sbjct: 329 ALRDDSVGCVRKTKLSCDGRDGFVRLKKMRLPDTTETSVDKGIGLKECEERCLKGCNCTA 388
Query: 337 F 337
F
Sbjct: 389 F 389
>At1g61610
Length = 842
Score = 56.2 bits (134), Expect = 3e-08
Identities = 62/235 (26%), Positives = 99/235 (41%), Gaps = 20/235 (8%)
Query: 29 ATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISV-NADYGKVVWVYDINH 87
+TS+S + L S+ F L F NS + + I N + VVWV +
Sbjct: 27 STSNSFTRNHTIREGDSLISEDESFELGFFTPKNSTLRYVGIWYKNIEPQTVVWVANREK 86
Query: 88 SIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLPNGSM 147
+ + L + G L I + + I + +NNTVA + G+ VL +
Sbjct: 87 PLLDHKGALKIADDGNLVIVNGQNETIWSTNVEPESNNTVAVLFKTGDLVLCS--DSDRR 144
Query: 148 SVLWQSFDYPSDVLIPMMKLGVNRKTGHN-----WSLVSD----KFNLEWEPKQG-ELNI 197
W+SF+ P+D +P M++ VN G N W SD K+++ +P E+ I
Sbjct: 145 KWYWESFNNPTDTFLPGMRVRVNPSLGENRAFIPWKSESDPSPGKYSMGIDPVGALEIVI 204
Query: 198 KKSGKVYWKSGKLKSNGLFENIPANVQSR---YQYIIVSNKDEDS---FTFEVKD 246
+ K W+SG S +F IP ++ Y + + S D D FT+ D
Sbjct: 205 WEGEKRKWRSGPWNS-AIFTGIPDMLRFTNYIYGFKLSSPPDRDGSVYFTYVASD 258
>At1g11280 serine/threonine kinase like protein
Length = 830
Score = 55.8 bits (133), Expect = 4e-08
Identities = 56/223 (25%), Positives = 93/223 (41%), Gaps = 22/223 (9%)
Query: 11 VLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFI 70
++++ W W + C A P L L S G + L F + +NS Q + I
Sbjct: 20 IVLFPWFLWLSLFLSCGYAAITISSP---LTLGQTLSSPGGFYELGFFSPNNSQNQYVGI 76
Query: 71 SVNADYGKVV-WVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVAT 129
+VV WV + I + L++ +G L + ++ + P +N A
Sbjct: 77 WFKKITPRVVVWVANREKPITTPVANLTISRNGSLILLDSSKNVVWSTRRPSISNKCHAK 136
Query: 130 MLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWE 189
+LD GN V+ + + S ++LWQSF+ P D ++P L N TG L S K + +
Sbjct: 137 LLDTGNLVI---VDDVSENLLWQSFENPGDTMLPYSSLMYNLATGEKRVLSSWKSHT--D 191
Query: 190 PKQGELNIK------------KSGKVYWKSGKLKSNGLFENIP 220
P G+ ++ + VY +SG G F +P
Sbjct: 192 PSPGDFVVRLTPQVPAQIVTMRGSSVYKRSGPWAKTG-FTGVP 233
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,765,699
Number of Sequences: 26719
Number of extensions: 458415
Number of successful extensions: 1267
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 1168
Number of HSP's gapped (non-prelim): 98
length of query: 381
length of database: 11,318,596
effective HSP length: 101
effective length of query: 280
effective length of database: 8,619,977
effective search space: 2413593560
effective search space used: 2413593560
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140024.11


