

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.4 - phase: 0
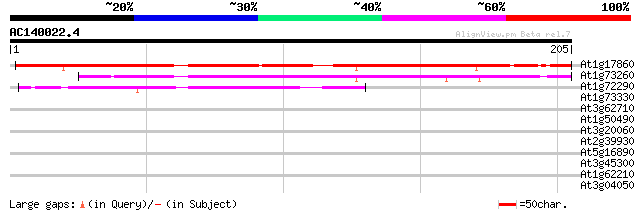
(205 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g17860 Unknown protein (F2H15.9) 166 6e-42
At1g73260 putative trypsin inhibitor (At1g73260) 103 8e-23
At1g72290 drought induced protein, putative 41 5e-04
At1g73330 unknown protein 33 0.080
At3g62710 beta-D-glucan exohydrolase-like protein 31 0.52
At1g50490 cyclin-specific ubiquitin carrier protein, putative 30 1.1
At3g20060 ubiquitin-conjugating enzyme, putative 29 1.5
At2g39930 putative isoamylase 29 2.0
At5g16890 unknown protein (At5g16890) 28 2.6
At3g45300 isovaleryl-CoA-dehydrogenase precursor (IVD) 27 5.7
At1g62210 unknown protein 27 7.5
At3g04050 putative pyruvate kinase 27 9.7
>At1g17860 Unknown protein (F2H15.9)
Length = 196
Score = 166 bits (421), Expect = 6e-42
Identities = 100/210 (47%), Positives = 131/210 (61%), Gaps = 23/210 (10%)
Query: 3 TSFLAFSIIFLAFICK----TFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNE 58
+S L ++ FI T AA EPV DI+GK + TGV YYILPVIRG+GGGLT+ N
Sbjct: 2 SSLLYIFLLLAVFISHRGVTTEAAVEPVKDINGKSLLTGVNYYILPVIRGRGGGLTMSNL 61
Query: 59 NNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQ 118
TCP V+Q++ EV G V F+PY+ K I STD+NIK T
Sbjct: 62 KT-----ETCPTSVIQDQFEVSQGLPVKFSPYD-KSRTIPVSTDVNIKFSPTS------- 108
Query: 119 VWKLNKV--LSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK-CQTLCR 175
+W+L + WF++T GVEGNPG T+ NWFKI+K +KDY FCP+VC C+ +CR
Sbjct: 109 IWELANFDETTKQWFISTCGVEGNPGQKTVDNWFKIDKFEKDYKIRFCPTVCNFCKVICR 168
Query: 176 ELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
++G++V D GK+ LALSD VP +V+FKRA
Sbjct: 169 DVGVFVQD-GKRRLALSD-VP-LKVMFKRA 195
>At1g73260 putative trypsin inhibitor (At1g73260)
Length = 215
Score = 103 bits (256), Expect = 8e-23
Identities = 66/184 (35%), Positives = 100/184 (53%), Gaps = 12/184 (6%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
V+DI G + YY+LPVIRG+GGGLT+ CP ++QE EV G V
Sbjct: 30 VVDIDGNAMFHE-SYYVLPVIRGRGGGLTLAGRGG-----QPCPYDIVQESSEVDEGIPV 83
Query: 86 TFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPGF 143
F+ + K + S +LNI++ V T C QS W++ + +F+ G G
Sbjct: 84 KFSNWRLKVAFVPESQNLNIETDVGATICIQSTYWRVGEFDHERKQYFVVAGPKPEGFGQ 143
Query: 144 DTIFNWFKIEKADKD-YVFSFCPSVCKC-QTLCRELGLYVYDHGKKHLALSDQVPSFRVV 201
D++ ++FKIEK+ +D Y F FCP C C ++G+++ + G + LALSD+ F V+
Sbjct: 144 DSLKSFFKIEKSGEDAYKFVFCPRTCDSGNPKCSDVGIFIDELGVRRLALSDK--PFLVM 201
Query: 202 FKRA 205
FK+A
Sbjct: 202 FKKA 205
>At1g72290 drought induced protein, putative
Length = 215
Score = 40.8 bits (94), Expect = 5e-04
Identities = 39/129 (30%), Positives = 55/129 (42%), Gaps = 17/129 (13%)
Query: 4 SFLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPV--IRGKGGGLTVVNENNL 61
SFL ++F A IC EPV D +G + T +Y+I PV GGGL L
Sbjct: 8 SFLII-LLFAATICTH--GNEPVKDTAGNPLNTREQYFIQPVKTESKNGGGLVPAAITVL 64
Query: 62 NGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWK 121
CPL + Q L + G V+F ++TS+ +NI+ +S +W
Sbjct: 65 ----PFCPLGITQTLLPYQPGLPVSFVLALGVGSTVMTSSAVNIE--------FKSNIWP 112
Query: 122 LNKVLSGVW 130
K S W
Sbjct: 113 FCKEFSKFW 121
>At1g73330 unknown protein
Length = 209
Score = 33.5 bits (75), Expect = 0.080
Identities = 46/208 (22%), Positives = 89/208 (42%), Gaps = 28/208 (13%)
Query: 6 LAFSIIFL--AFICKTFAAPEP-VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLN 62
++ + IFL A + A P+ V D G+ + G Y+I+P GGG+ +
Sbjct: 5 ISITTIFLVVALAAPSLARPDNHVEDSVGRLLRPGQTYHIVPANPETGGGI-------FS 57
Query: 63 GNNNTCPLYVLQEKLEVKNGQAVTFTP--YNAKKGVILTSTDLNIKSYVTKTTCAQSQVW 120
+ CPL + Q + G + F + K+ + S + ++ +S+ W
Sbjct: 58 NSEEICPLDIFQSNNPLDLGLPIKFKSELWFVKE---MNSITIEFEAPNWFLCPKESKGW 114
Query: 121 KL--NKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKD-YVFSFCPSVCKCQTLCREL 177
++ ++ ++TGG NP + F+I + D Y +C ++ T C +
Sbjct: 115 RVVYSEEFKKSLIISTGG-SSNP------SGFQIHRVDGGAYKIVYCTNI--STTTCMNV 165
Query: 178 GLYVYDHGKKHLALSDQVPSFRVVFKRA 205
G++ G + LAL+ + V F++A
Sbjct: 166 GIFTDISGARRLALTSD-EALLVKFQKA 192
>At3g62710 beta-D-glucan exohydrolase-like protein
Length = 650
Score = 30.8 bits (68), Expect = 0.52
Identities = 19/66 (28%), Positives = 32/66 (47%), Gaps = 1/66 (1%)
Query: 48 GKGGGLTVVNENNLNGNNNTC-PLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIK 106
G GG + +NENN +N T +++ ++ VK G A Y++ GV + + I
Sbjct: 244 GDGGTINGINENNTVADNATLFGIHMPPFEIAVKKGIASIMASYSSLNGVKMHANRAMIT 303
Query: 107 SYVTKT 112
Y+ T
Sbjct: 304 DYLKNT 309
>At1g50490 cyclin-specific ubiquitin carrier protein, putative
Length = 180
Score = 29.6 bits (65), Expect = 1.1
Identities = 25/88 (28%), Positives = 32/88 (35%), Gaps = 15/88 (17%)
Query: 110 TKTTCAQSQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFS------- 162
TKT +QS + +L L G+ G+ P D IF W KD VF
Sbjct: 28 TKTVDSQSVLKRLQSELMGLMMGGGPGISAFPEEDNIFCWKGTITGSKDTVFEGTEYRLS 87
Query: 163 --------FCPSVCKCQTLCRELGLYVY 182
F P K +T C + VY
Sbjct: 88 LSFSNDYPFKPPKVKFETCCFHPNVDVY 115
>At3g20060 ubiquitin-conjugating enzyme, putative
Length = 181
Score = 29.3 bits (64), Expect = 1.5
Identities = 18/52 (34%), Positives = 23/52 (43%)
Query: 110 TKTTCAQSQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVF 161
TKT + S + +L L G+ A G+ P D IF W KD VF
Sbjct: 29 TKTVDSHSVLKRLQSELMGLMMGADPGISAFPEEDNIFCWKGTITGSKDTVF 80
>At2g39930 putative isoamylase
Length = 783
Score = 28.9 bits (63), Expect = 2.0
Identities = 15/41 (36%), Positives = 21/41 (50%), Gaps = 1/41 (2%)
Query: 147 FNWFKIEKADKDYVFSFCPSVCKCQTLCRELGLYVYDHGKK 187
F W K E+A D+ F FC + K + C LGL + K+
Sbjct: 633 FRWDKKEEAHSDF-FRFCRILIKFRDECESLGLNDFPTAKR 672
>At5g16890 unknown protein (At5g16890)
Length = 511
Score = 28.5 bits (62), Expect = 2.6
Identities = 15/52 (28%), Positives = 24/52 (45%), Gaps = 10/52 (19%)
Query: 125 VLSGVWFLATGGVEGNPGFDTIFNWFKIEKA--DKDYVFSFCPSVCKCQTLC 174
V + +W L P D+ NW+K + +KD + + P+V C T C
Sbjct: 255 VKNAIWLL--------PDMDSTGNWYKPGQVSLEKDLILPYVPNVDICDTKC 298
>At3g45300 isovaleryl-CoA-dehydrogenase precursor (IVD)
Length = 409
Score = 27.3 bits (59), Expect = 5.7
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 1/62 (1%)
Query: 101 TDLNIKSYVTKTTCAQSQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYV 160
++L I V T AQ + + L K++SG A E N G D + K EK D Y+
Sbjct: 119 SNLCINQLVRNGTAAQKEKY-LPKLISGEHVGALAMSEPNAGSDVVGMKCKAEKVDGGYI 177
Query: 161 FS 162
+
Sbjct: 178 LN 179
>At1g62210 unknown protein
Length = 176
Score = 26.9 bits (58), Expect = 7.5
Identities = 24/83 (28%), Positives = 33/83 (38%), Gaps = 13/83 (15%)
Query: 35 TTGVKYYILPVIRG----KGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPY 90
T G K ++ VI G K G T+ + LNG PL + G+A T
Sbjct: 77 TNGTKTPVIGVIVGVSPIKESGATIGTKAPLNGVLRPAPL---------REGEATLATNN 127
Query: 91 NAKKGVILTSTDLNIKSYVTKTT 113
K V + N++S V TT
Sbjct: 128 TGKPSVSVMGVGTNVQSEVLFTT 150
>At3g04050 putative pyruvate kinase
Length = 510
Score = 26.6 bits (57), Expect = 9.7
Identities = 14/60 (23%), Positives = 30/60 (49%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
+LD G ++ TG PV +G +T+ + + G++NT + + ++K+G +
Sbjct: 78 MLDTKGPEIRTGFLKEGKPVELIQGQEITISTDYTMEGDSNTISMSYKKLAEDLKSGDVI 137
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,919,273
Number of Sequences: 26719
Number of extensions: 212355
Number of successful extensions: 447
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 436
Number of HSP's gapped (non-prelim): 12
length of query: 205
length of database: 11,318,596
effective HSP length: 95
effective length of query: 110
effective length of database: 8,780,291
effective search space: 965832010
effective search space used: 965832010
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC140022.4


