

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139854.8 - phase: 0
(304 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
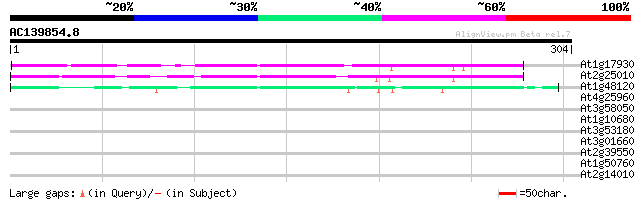
Score E
Sequences producing significant alignments: (bits) Value
At1g17930 unknown protein 86 2e-17
At2g25010 unknown protein 72 5e-13
At1g48120 serine/threonine phosphatase PP7, putative 65 5e-11
At4g25960 P-glycoprotein-2 (pgp2) 29 2.8
At3g58050 putative protein 29 2.8
At1g10680 putative P-glycoprotein-2 emb|CAA71277 29 2.8
At3g53180 nodulin / glutamate-ammonia ligase - like protein 29 3.6
At3g01660 hypothetical protein 28 4.7
At2g39550 putative geranylgeranyl transferase type I beta subunit 28 4.7
At1g50760 hypothetical protein 28 4.7
At2g14010 hypothetical protein 28 6.2
>At1g17930 unknown protein
Length = 478
Score = 85.9 bits (211), Expect = 2e-17
Identities = 73/284 (25%), Positives = 128/284 (44%), Gaps = 33/284 (11%)
Query: 2 PMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKYG 61
P GEMT+TLD+V+ + L ++G+ + K+ K E +Q+ L + E N+
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVVGVKE--KDEDPSQVCLRLLGKLPKGELSGNR--- 138
Query: 62 GYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSN 121
++ L++ + A + +E+E Y R YL+Y+VG +F
Sbjct: 139 --VTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTDP 182
Query: 122 KRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFLAH 181
+I + YL D + Y+WG LA+LY ++ +A + +GG +TLL W H
Sbjct: 183 SKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQCWSYFH 241
Query: 182 FPGFFSVDLNTDYLENYPVAARWK---LPKGHGEGITYWSLLDRIQLDDVCYKPYKEHRE 238
++D +P+A WK + + Y LD + +V + P++ +
Sbjct: 242 ----LNIDRPKRTTRQFPLALLWKGRQQSRSKNDLFKYRKALDDLDPSNVSWCPFEGDLD 297
Query: 239 I--QDFEE--VFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
I Q F++ + S + G + V H P R ++Q+G Q IP
Sbjct: 298 IVPQSFKDNLLLGRSRTKLIGPKVVEWHFPDRCMKQFGLCQVIP 341
>At2g25010 unknown protein
Length = 509
Score = 71.6 bits (174), Expect = 5e-13
Identities = 73/287 (25%), Positives = 117/287 (40%), Gaps = 35/287 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P+GEMT+TLD+VA + L I+G + G K+ LG A K N
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPIV-GSKVGDEVAMDMCGRLLGKLPSAANKEVN--- 147
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEELE-ELARVRTYCVRCYLLYLVGCLLFGDR 119
S +L ++ +E PE+ ++ + T R YLLYL+G +F
Sbjct: 148 ---CSRVKLNWLKRTF---------SECPEDASFDVVKCHT---RAYLLYLIGSTIFATT 192
Query: 120 SNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTWFL 179
++ + YL D + Y+WG LA LY L +A + G +TLL W
Sbjct: 193 DGDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQCW-- 249
Query: 180 AHFPGFFSVDLNTDYLEN--YPVAARW--KLPKGHGEGITYWSLLDRIQLDDVCYKPYKE 235
+F +D+ +P+A W K + + Y LD + + + PY+
Sbjct: 250 ----SYFHLDIGRPEKSEACFPLALLWKGKGSRSKTDLSEYRRELDDLDPSKITWCPYER 305
Query: 236 HREI----QDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYVQTIP 278
+ + + S + ++ H P R LRQ+G Q IP
Sbjct: 306 FENLIPPHIKAKLILGRSKTTLVCFEKIELHFPDRCLRQFGKRQPIP 352
>At1g48120 serine/threonine phosphatase PP7, putative
Length = 1338
Score = 65.1 bits (157), Expect = 5e-11
Identities = 86/323 (26%), Positives = 130/323 (39%), Gaps = 62/323 (19%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAAQLMTYLGVSQHEAEKICNQKY 60
+P GE+TVTL DV L L ++G + G +++ +C
Sbjct: 101 LPAGEITVTLQDVNILLGLRVDGPAVT------------------GSTKYNWADLCEDLL 142
Query: 61 GGYISYPRLRDFYTSYLG----RANVLAGTEDPEELEELARVRTYC-VRCYLLYLVGCLL 115
G P +D + S++ R N DP+E V C R ++L L+ L
Sbjct: 143 G---HRPGPKDLHGSHVSLAWLRENFRNLPADPDE------VTLKCHTRAFVLALMSGFL 193
Query: 116 FGDRSNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLT 175
+GD+S + L +L + D + + SWG TLA LY EL A + + G + LL
Sbjct: 194 YGDKSKHDVALTFLPLLRD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQ 252
Query: 176 TWFLAHF----PGFFSVDLNTDYLENY------PVAARWKL-----PKGHGEGITYW-SL 219
W PG D+ Y++ P+ R L P+G G+ ++
Sbjct: 253 LWAWERLHVGRPGRLK-DVGASYMDGIDGPLPDPLGCRASLSHKENPRG---GLDFYRDQ 308
Query: 220 LDRIQLDDVCYKPY-----KEHREIQDFEEVFWYSGWIMCGVRRVYRHLPKRVLRQYGYV 274
D+ + + V ++PY + I E W + + V H P RVLRQ+G
Sbjct: 309 FDQQKDEQVIWQPYTPDLLAKIPLICVSGENIWRTVAPLICFDVVEWHRPDRVLRQFGLH 368
Query: 275 QTIPRHPTDVRDLPTAFHCADVR 297
QTIP P D A H D R
Sbjct: 369 QTIPA-PCDNE---KALHAIDKR 387
>At4g25960 P-glycoprotein-2 (pgp2)
Length = 1233
Score = 29.3 bits (64), Expect = 2.8
Identities = 25/71 (35%), Positives = 35/71 (49%), Gaps = 14/71 (19%)
Query: 41 LMTY-LGVSQHEAEKICNQKYGGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVR 99
L TY L +S H +EK+ Q YGG D +YL +AN+LAG E + + V
Sbjct: 819 LATYPLVISGHISEKLFMQGYGG--------DLNKAYL-KANMLAG----ESVSNIRTVA 865
Query: 100 TYCVRCYLLYL 110
+C +L L
Sbjct: 866 AFCAEEKILEL 876
>At3g58050 putative protein
Length = 1209
Score = 29.3 bits (64), Expect = 2.8
Identities = 18/51 (35%), Positives = 25/51 (48%)
Query: 64 ISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCL 114
+SY +L+ F++ +A D + L E AR YC RC L L G L
Sbjct: 24 VSYNQLQKFWSELSPKARQELLKIDKQTLFEQARKNMYCSRCNGLLLEGFL 74
>At1g10680 putative P-glycoprotein-2 emb|CAA71277
Length = 1227
Score = 29.3 bits (64), Expect = 2.8
Identities = 25/66 (37%), Positives = 34/66 (50%), Gaps = 17/66 (25%)
Query: 41 LMTY-LGVSQHEAEKICNQKYGGYISYPRLRDFYTSYLGRANVLAGTEDPEELEELARVR 99
L TY L +S H +EKI Q YGG +S +YL +AN+LAG E ++ +R
Sbjct: 811 LATYPLIISGHISEKIFMQGYGGNLS--------KAYL-KANMLAG-------ESISNIR 854
Query: 100 TYCVRC 105
T C
Sbjct: 855 TVVAFC 860
>At3g53180 nodulin / glutamate-ammonia ligase - like protein
Length = 845
Score = 28.9 bits (63), Expect = 3.6
Identities = 17/61 (27%), Positives = 29/61 (46%), Gaps = 1/61 (1%)
Query: 134 DGYAGMRNYSWGGMTLA-YLYGELADACRPGHRALGGSVTLLTTWFLAHFPGFFSVDLNT 192
DGYA Y G ++ L+DAC G +L ++ F + GF+ ++++T
Sbjct: 328 DGYASPETYYLGAKKAREVIFLVLSDACASGDLSLMEAIDAAKDIFSRNSIGFYKLNIDT 387
Query: 193 D 193
D
Sbjct: 388 D 388
>At3g01660 hypothetical protein
Length = 273
Score = 28.5 bits (62), Expect = 4.7
Identities = 23/88 (26%), Positives = 39/88 (44%), Gaps = 5/88 (5%)
Query: 183 PGFFSVDLNTDYLENYPVAARWKLPKGHGEGITYWSLLDRIQLDDVCYKPYKEHREIQDF 242
PG +DL + ++ + P +++ GH G+ L +LD Y K+ E Q F
Sbjct: 94 PGSTVLDLMSSWVSHLPEEVKYEKVVGH--GLNAQELARNPRLD---YFFVKDLNEDQKF 148
Query: 243 EEVFWYSGWIMCGVRRVYRHLPKRVLRQ 270
E Y ++C V Y P++V +
Sbjct: 149 EFEDKYFDAVLCSVGVQYLQQPEKVFAE 176
>At2g39550 putative geranylgeranyl transferase type I beta subunit
Length = 375
Score = 28.5 bits (62), Expect = 4.7
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 1/24 (4%)
Query: 56 CNQKYGGYISYP-RLRDFYTSYLG 78
C KYGG+ +P +L D Y SY G
Sbjct: 323 CQSKYGGFSKFPGQLPDLYHSYYG 346
>At1g50760 hypothetical protein
Length = 649
Score = 28.5 bits (62), Expect = 4.7
Identities = 25/117 (21%), Positives = 43/117 (36%), Gaps = 28/117 (23%)
Query: 195 LENYPVAARWKLPKGHGEGITYWSLLDRIQLDDVCYKPYKEHREIQDFEEVFWYSGWIMC 254
L P A W K + +LD ++D ++PY R + +++ +Y MC
Sbjct: 276 LAGKPRVAIWNKLKQQTSNVR--QILDNSKIDSFEWRPYT--RSVTNWKFPQFYPEKAMC 331
Query: 255 ------------------------GVRRVYRHLPKRVLRQYGYVQTIPRHPTDVRDL 287
G+ +V + P RV Q+G +Q +P D +L
Sbjct: 332 VHVSPSLDEELISFARCIKDSELVGIEKVEHYFPNRVASQFGMLQDVPSPHVDRNNL 388
>At2g14010 hypothetical protein
Length = 833
Score = 28.1 bits (61), Expect = 6.2
Identities = 24/86 (27%), Positives = 36/86 (40%), Gaps = 3/86 (3%)
Query: 118 DRSNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHRALGGSVTLLTTW 177
D+ +KR+E L +AD A + + ++Y E+ RPG GGSV L T
Sbjct: 360 DKKDKRMESRMLKAIADSEARLSSLLRATHGKHHMYEEVQ---RPGLGGKGGSVVDLDTS 416
Query: 178 FLAHFPGFFSVDLNTDYLENYPVAAR 203
H + D+ L Y + R
Sbjct: 417 MGVHTMKNSATDILDQVLGEYTLVQR 442
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.142 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,509,081
Number of Sequences: 26719
Number of extensions: 332979
Number of successful extensions: 783
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 768
Number of HSP's gapped (non-prelim): 14
length of query: 304
length of database: 11,318,596
effective HSP length: 99
effective length of query: 205
effective length of database: 8,673,415
effective search space: 1778050075
effective search space used: 1778050075
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139854.8


