

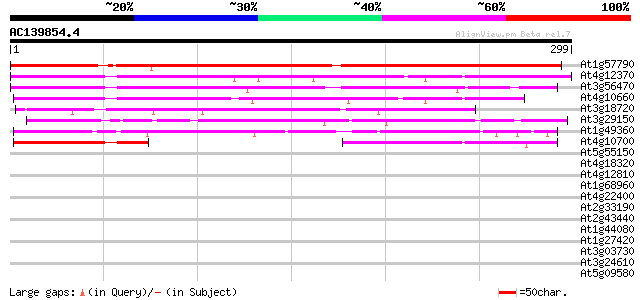
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139854.4 + phase: 0
(299 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g57790 unknown protein 243 9e-65
At4g12370 putative protein 219 2e-57
At3g56470 putative protein 199 2e-51
At4g10660 hypothetical protein 198 3e-51
At3g18720 unknown protein 139 2e-33
At3g29150 hypothetical protein 128 4e-30
At1g49360 unknown protein 127 7e-30
At4g10700 hypothetical protein 87 1e-17
At5g55150 putative protein 38 0.006
At4g18320 hypothetical protein 37 0.010
At4g12810 putative protein 36 0.029
At1g68960 hypothetical protein 34 0.11
At4g22400 putative protein 33 0.24
At2g33190 hypothetical protein 33 0.24
At2g43440 hypothetical protein 32 0.42
At1g44080 hypothetical protein 32 0.42
At1g27420 hypothetical protein 32 0.42
At3g03730 hypothetical protein 32 0.54
At3g24610 hypothetical protein 31 0.93
At5g09580 putative protein 30 1.2
>At1g57790 unknown protein
Length = 352
Score = 243 bits (620), Expect = 9e-65
Identities = 129/298 (43%), Positives = 186/298 (62%), Gaps = 14/298 (4%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPE-LDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSL 59
MYFP+ N YDFYDP K Y++ELP+ L G V Y+KDGWLL++++D +H F L
Sbjct: 60 MYFPETKNTYDFYDPSNCKKYTMELPKSLVGFIVRYSKDGWLLMSQED-----SSH-FVL 113
Query: 60 FNPFTRDLITLPKFD--RTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWT 117
FNPFT D++ LP YQ+ FS APTS+ CV+ + V I T PG+ WT
Sbjct: 114 FNPFTMDVVALPFLHLFTYYQLVGFSSAPTSSECVVFTIKDYDPGHVTIRTWSPGQTMWT 173
Query: 118 TVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
++ +++ + +VFSNG+FYCL+ R + VFDP RTW V VPPP+C + K
Sbjct: 174 SMQVESQFLDVDHNNVVFSNGVFYCLNQRNHVAVFDPSLRTWNVLDVPPPRCPDD----K 229
Query: 178 NWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRT 237
+W++GKFM+ +KG+I V+ +DP++FKLDLT W+E +L +T+F S S SRT
Sbjct: 230 SWNEGKFMVGYKGDILVIRTYENKDPLVFKLDLTRGIWEEKDTLGSLTIFVSRKSCESRT 289
Query: 238 YA-TGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPPKD 294
Y G++RNSVYFP++ + K+ + +S D+ RY+ E D + + +N WIEPPK+
Sbjct: 290 YVKDGMLRNSVYFPELCYNEKQSVVYSFDEGRYHLREHDLDWGKQLSSDNIWIEPPKN 347
>At4g12370 putative protein
Length = 300
Score = 219 bits (557), Expect = 2e-57
Identities = 122/309 (39%), Positives = 172/309 (55%), Gaps = 19/309 (6%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
MY PK+GN ++ YDP+ +K Y+L LPEL VCY++DGWLL+ + R + F
Sbjct: 1 MYLPKRGNLFELYDPLHQKMYTLNLPELAKSTVCYSRDGWLLMRKTISREM------FFF 54
Query: 61 NPFTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTT-- 118
NPFTR+LI +PK +Y AFSCAPTS CV+L F+ V + STC+P EW T
Sbjct: 55 NPFTRELINVPKCTLSYDAIAFSCAPTSGTCVLLAFKHVSYRITTTSTCHPKATEWVTED 114
Query: 119 VYYDAELSCSMCD--KLVFSNGLFYCLSDRGWLGVFDPLERTWT---VFKVPPPKCLAES 173
+ + + +V++ FYCL +G L FDP R W + +P P
Sbjct: 115 LQFHRRFRSETLNHSNVVYAKRRFYCLDGQGSLYYFDPSSRRWDFSYTYLLPCPYISDRF 174
Query: 174 STAKNWSKGK-FMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRS--LNGVTLFASF 230
S K + F+ KG F + C GE PI+ KL+ + W+E+ S ++G+T+F
Sbjct: 175 SYQYERKKKRIFLAVRKGVFFKIFTCDGEKPIVHKLED--INWEEINSTTIDGLTIFTGL 232
Query: 231 LSSHSRTYATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIE 290
SS R MRNSVYFP++RF KRC+S+SLD+ RYYP +Q +++ + EN WI
Sbjct: 233 YSSEVRLNLPW-MRNSVYFPRLRFNVKRCVSYSLDEERYYPRKQWQEQEDLCPIENLWIR 291
Query: 291 PPKDFTGWM 299
PPK +M
Sbjct: 292 PPKKAVDFM 300
>At3g56470 putative protein
Length = 367
Score = 199 bits (505), Expect = 2e-51
Identities = 115/298 (38%), Positives = 163/298 (54%), Gaps = 25/298 (8%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
+YF K + Y+ YDP +K +L PEL G RVCY+KDGWLL+ + +L F
Sbjct: 76 IYFSKTDDSYELYDPSMQKNCNLHFPELSGFRVCYSKDGWLLMYNPNSYQL------LFF 129
Query: 61 NPFTRDLITLPKFDRTY-QIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTV 119
NPFTRD + +P Y Q AFSCAPTST C++ V + + I T + KEW T
Sbjct: 130 NPFTRDCVPMPTLWMAYDQRMAFSCAPTSTSCLLFTVTSVTWNYITIKTYFANAKEWKTS 189
Query: 120 YYDAEL--SCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
+ L + + +++VFSNG+FYCL++ G L +FDP W V PPK +
Sbjct: 190 VFKNRLQRNFNTFEQIVFSNGVFYCLTNTGCLALFDPSLNYWNVLPGRPPK--------R 241
Query: 178 NWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRT 237
S G FM EH+G IF++++ +P + KLDLT EW E ++L G+T++AS LSS SR
Sbjct: 242 PGSNGCFMTEHQGEIFLIYMYRHMNPTVLKLDLTSFEWAERKTLGGLTIYASALSSESRA 301
Query: 238 ---YATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPP 292
+GI N + + + CI + +D+ SE C + N +EN WI PP
Sbjct: 302 EQQKQSGIW-NCLCLSVFHGFKRTCIYYKVDEE----SEVCFKWKKQNPYENIWIMPP 354
>At4g10660 hypothetical protein
Length = 317
Score = 198 bits (504), Expect = 3e-51
Identities = 117/283 (41%), Positives = 155/283 (54%), Gaps = 24/283 (8%)
Query: 3 FPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNP 62
FP + +DP++RK Y+L LPEL G VCY+KDGWLL+ R + FNP
Sbjct: 24 FPNHEDLTFLFDPLERKRYTLNLPELVGTDVCYSKDGWLLMRRSSLVDM------FFFNP 77
Query: 63 FTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTVYYD 122
+TR+LI LPK + +Q AFS APTS CV+L R ++ IS CYPG EW T
Sbjct: 78 YTRELINLPKCELAFQAIAFSSAPTSGTCVVLALRPFTRYIIRISICYPGATEWIT---- 133
Query: 123 AELSCS------MCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTA 176
E SCS M LV++N FYC S G L FD RT + +C +
Sbjct: 134 QEFSCSLRFDPYMHSNLVYANDHFYCFSSGGVLVDFDVASRTMSEQAWNEHRCAYMRNDN 193
Query: 177 KNW---SKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRS--LNGVTLFASFL 231
W K ++ E KG +F+++ C E P+++K L W+E+ S L+GVT+FAS
Sbjct: 194 AEWFNLPKRIYLAEQKGELFLMYTCSSEIPMVYK--LVSSNWEEINSTTLDGVTIFASMY 251
Query: 232 SSHSRTYATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQ 274
SS +R G MRNSVYFPK K C+S+S D+ RYYP +Q
Sbjct: 252 SSETRLDVLG-MRNSVYFPKYGLDCKGCVSYSFDEARYYPRKQ 293
>At3g18720 unknown protein
Length = 380
Score = 139 bits (349), Expect = 2e-33
Identities = 90/253 (35%), Positives = 133/253 (51%), Gaps = 18/253 (7%)
Query: 4 PKKGNCYDFYDPVQRKTYSLELPELDGCR--VCYTKDGWLLLNRQDWRRLDGNHIFSLFN 61
PK+G+ Y ++P + +T+ L+ PEL G R + KDGWLL+ + D + N
Sbjct: 105 PKEGD-YVLFNPSRSQTHHLKFPELTGYRNKLACAKDGWLLVVK------DNPDVVFFLN 157
Query: 62 PFTRDLITLPKFDR--TYQIAAFSCAPTSTGCVILIFRRVGS--SLVAISTCYPGEKEWT 117
PFT + I LP+ + T FS APTST C ++ F ++V + T PGE WT
Sbjct: 158 PFTGERICLPQVPQNSTRDCLTFSAAPTSTSCCVISFTPQSFLYAVVKVDTWRPGESVWT 217
Query: 118 TVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
T ++D + + ++ +FSNG+FYCLS G L VFDP TW V V P +
Sbjct: 218 THHFDQKRYGEVINRCIFSNGMFYCLSTSGRLSVFDPSRETWNVLPVKPCRAFRRKIML- 276
Query: 178 NWSKGKFMIEHKGNIFVV--HICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHS 235
+ FM EH+G+IFVV + FKL+L W+E++ NG+T+F+S +S +
Sbjct: 277 --VRQVFMTEHEGDIFVVTTRRVNNRKLLAFKLNLQGNVWEEMKVPNGLTVFSSDATSLT 334
Query: 236 RTYATGIMRNSVY 248
R RN +Y
Sbjct: 335 RAGLPEEERNILY 347
>At3g29150 hypothetical protein
Length = 417
Score = 128 bits (321), Expect = 4e-30
Identities = 90/295 (30%), Positives = 143/295 (47%), Gaps = 25/295 (8%)
Query: 10 YDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNPFTRDLIT 69
Y +DP++ ++Y E P L + K+GWLL+++ D+ +F L NPFTR+ +
Sbjct: 111 YILFDPLRNQSYKQEFPGLGCHKFLSYKNGWLLVSKYDYP----GALFFL-NPFTRERVY 165
Query: 70 LPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTVYYDAELSCSM 129
LP AFS APTST C++ + I T GE WTT + +
Sbjct: 166 LPLLVPC--CGAFSAAPTSTSCLVACV----NYYFEIMTWRLGETVWTTNCFGNHIRGRK 219
Query: 130 CDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPP-----PKCLAESSTAKNWSKGKF 184
DK VFSNG+F+CLS G+L VFDP TW + V P CLA + +
Sbjct: 220 WDKCVFSNGMFFCLSTCGYLRVFDPHRATWNILPVKPCILPLEPCLAFRQPSLMGMR-VL 278
Query: 185 MIEHKGNIFVVHICC--GEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRTYATGI 242
M+EH+G+++V+ +FKL+L W+E + L G+T+FA + +S +R +
Sbjct: 279 MMEHEGDVYVISTFSNYNNQASVFKLNLKREVWEEKKELGGLTVFAGYHTSLTRASLLAL 338
Query: 243 MRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPPKDFTG 297
RN++Y + + +SL ++ +C + W++PP + G
Sbjct: 339 DRNNMY---TSHAARSFVYYSLAGNKF---SRCPPGCNYLSDRFAWVDPPHNQAG 387
>At1g49360 unknown protein
Length = 481
Score = 127 bits (319), Expect = 7e-30
Identities = 105/314 (33%), Positives = 149/314 (47%), Gaps = 40/314 (12%)
Query: 3 FPKKGNCYDFYDPVQRK-TYSLELPELD-GCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
F KG Y F+DPV++K T + LPEL + Y+KDGWLL+N D L + F F
Sbjct: 160 FQDKGVSYGFFDPVEKKKTKEMNLPELSKSSGILYSKDGWLLMN--DSLSLIADMYF--F 215
Query: 61 NPFTRDLITLPK---FDRTYQIAAFSCAPTSTGCVILIFRRVGSSL-VAISTCYPGEKEW 116
NPFTR+ I LP+ + + AFSCAPT C++ + SS+ + IST PG W
Sbjct: 216 NPFTRERIDLPRNRIMESVHTNFAFSCAPTKKSCLVFGINNISSSVAIKISTWRPGATTW 275
Query: 117 TTVYYDAELSCSM--CDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESS 174
+ +++S+GLFY S+ LGVFDP RTW V V P S
Sbjct: 276 LHEDFPNLFPSYFRRLGNILYSDGLFYTASETA-LGVFDPTARTWNVLPVQPIPMAPRSI 334
Query: 175 TAKNWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSH 234
++M E++G+IF+V +P++++L+ W++ +L+G ++F S S
Sbjct: 335 --------RWMTEYEGHIFLVD-ASSLEPMVYRLNRLESVWEKKETLDGSSIFLSDGSCV 385
Query: 235 SRTYATGIMRNSVYFPKVRFYGKR------CISFSLDDRRY--YPSEQCRDKVEPNTFE- 285
TG M N +YF RF +R C +Y Y C D E FE
Sbjct: 386 MTYGLTGSMSNILYFWS-RFINERRSTKSPCPFSRNHPYKYSLYSRSSCEDP-EGYYFEY 443
Query: 286 -------NFWIEPP 292
WIEPP
Sbjct: 444 LTWGQKVGVWIEPP 457
>At4g10700 hypothetical protein
Length = 247
Score = 87.0 bits (214), Expect = 1e-17
Identities = 48/121 (39%), Positives = 70/121 (57%), Gaps = 7/121 (5%)
Query: 178 NWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRT 237
N K ++ E KG +F+++ C E P+++KL + +E +L+GVT+FAS SS +R
Sbjct: 119 NLPKRIYLAEQKGELFLMYTCSSEIPMVYKLVSSNLEEMNSTTLDGVTIFASMYSSETRL 178
Query: 238 YATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQ------CRDKVEPNTFENFWIEP 291
G MRNSVYFPK K C+S+S D+ RYYP +Q + + E + WIEP
Sbjct: 179 DVLG-MRNSVYFPKYGLDCKGCVSYSFDEARYYPRKQFPKPTMWQMQKELCPLRSLWIEP 237
Query: 292 P 292
P
Sbjct: 238 P 238
Score = 70.9 bits (172), Expect = 8e-13
Identities = 34/72 (47%), Positives = 45/72 (62%), Gaps = 6/72 (8%)
Query: 3 FPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNP 62
FP + +DP++RK Y+L LPE+ G VCY+KDGWLL+ R + FNP
Sbjct: 24 FPNHEDMTFLFDPLERKRYTLNLPEVVGTDVCYSKDGWLLMRRSSLVDM------FFFNP 77
Query: 63 FTRDLITLPKFD 74
+TR+LI LPKFD
Sbjct: 78 YTRELINLPKFD 89
>At5g55150 putative protein
Length = 340
Score = 38.1 bits (87), Expect = 0.006
Identities = 59/208 (28%), Positives = 88/208 (41%), Gaps = 40/208 (19%)
Query: 106 ISTCYPGEKEWTTVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVP 165
++ C G+K+WT D E S D +VF NG+F+ + DR LG E + K
Sbjct: 146 LAFCRRGDKQWT----DLESVASSVDDIVFCNGVFFAI-DR--LGEIYHCELSANNPKAT 198
Query: 166 PPKCLAESSTAKNWSKGKFMIEHK-GNIFVV-------HICCGEDPI-IFKLDLTLMEWK 216
P L +S + S K++ E ++VV C E I++ + EW
Sbjct: 199 P---LCSTSPFRYDSCKKYLAESDYDELWVVLKKLELNDDCDFETSFEIYEFNRETNEWT 255
Query: 217 EVRSLNGVTLFASFLSSHSRTYAT-----GIMR-NSVYF-----PKVRFYGKRCISFSLD 265
+V SL G L FLS R A G + NSVYF P V G + +S
Sbjct: 256 KVMSLRGKAL---FLSPQGRCIAVLAGERGFFKDNSVYFIDGDDPSVGGSGPQNLSVF-- 310
Query: 266 DRRYYPSEQCRDKVEPNTF--ENFWIEP 291
+ S+Q +P ++ + FW+ P
Sbjct: 311 ---EWESKQIMKIYQPRSWNCQMFWVTP 335
>At4g18320 hypothetical protein
Length = 320
Score = 37.4 bits (85), Expect = 0.010
Identities = 19/71 (26%), Positives = 34/71 (47%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
+Y + FYDPV+ + + + P L+G + + GW++++ +H L+
Sbjct: 69 LYRERGSRVAHFYDPVREELHHGKDPYLEGAKFLGSTLGWVVMSNSSAIHRRQDHQAFLY 128
Query: 61 NPFTRDLITLP 71
NPFT LP
Sbjct: 129 NPFTSQRYQLP 139
>At4g12810 putative protein
Length = 382
Score = 35.8 bits (81), Expect = 0.029
Identities = 62/267 (23%), Positives = 96/267 (35%), Gaps = 43/267 (16%)
Query: 5 KKGNCYDFYDPVQRKTYSLELPELD--GCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNP 62
KK +C YDP + KTY + D R + W L+ LD F L N
Sbjct: 83 KKDSC-KLYDPHENKTYIVRDLGFDLVTSRCLASSGSWFLM-------LDHRTEFHLLNL 134
Query: 63 FTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVA--ISTCY----PGEKEW 116
FTR I LP + T V+ + + LV IS+ + G+ W
Sbjct: 135 FTRVRIPLPSLESTR-----GSDIKIGNAVLWVDEQRKDYLVVWNISSLFGYHKKGDDRW 189
Query: 117 TTVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTA 176
C + +VF Y LS G + VF + P +C S+
Sbjct: 190 KVFKPLENERCIIA--MVFKENKLYVLSVDGNVDVF------YFSGNDSPVRCATLPSSP 241
Query: 177 KNWSKG-KFMIEHKGNIFVVHICCGEDP-------IIFKLDLTLMEWKEVRSLNGVTLFA 228
KG K ++ G + ++ P ++K+D W+ ++SL G L
Sbjct: 242 LR--KGHKVVVTLSGEVLIIVAKVEPYPRTRLCFFAVYKMDPKSSRWETIKSLAGEALIL 299
Query: 229 SFLSSHSRTYATGIMRNSVYFPKVRFY 255
T +M+N +YF +F+
Sbjct: 300 DL----GITVEAKVMKNCIYFSNDQFH 322
>At1g68960 hypothetical protein
Length = 376
Score = 33.9 bits (76), Expect = 0.11
Identities = 30/118 (25%), Positives = 58/118 (48%), Gaps = 17/118 (14%)
Query: 10 YDFYDPVQRKTYSLE----LPELDGCRVCYTKDGWL-LLNRQDWRRLDGNHIFSLFNPFT 64
++ YDP +++ +E E+ + GWL LLN++D + ++ NP +
Sbjct: 57 FNLYDPRKQEQVKIENKILSKEVYESHKLGSSRGWLALLNKKD----STVRLTNILNP-S 111
Query: 65 RDLITLPKFDRTYQIAAFSCAPTSTG---CVILIFRRVGSSLVAISTCYPGEKEWTTV 119
+ +I+LP R + + + +S+ CV+ + + GS +S C PG+ EWT +
Sbjct: 112 KKIISLPSLTRDKYESHVNVSVSSSNEEDCVVAV-KFYGSR---VSLCRPGDSEWTRI 165
>At4g22400 putative protein
Length = 327
Score = 32.7 bits (73), Expect = 0.24
Identities = 18/61 (29%), Positives = 29/61 (47%)
Query: 12 FYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNPFTRDLITLP 71
F+DPV+ + + P L R + GWL+++ H L+NPF +L LP
Sbjct: 80 FFDPVRERVHRGNDPRLADARFLGSTLGWLVMSESLDLNPRKTHQTFLYNPFISELQQLP 139
Query: 72 K 72
+
Sbjct: 140 E 140
>At2g33190 hypothetical protein
Length = 379
Score = 32.7 bits (73), Expect = 0.24
Identities = 50/250 (20%), Positives = 93/250 (37%), Gaps = 55/250 (22%)
Query: 13 YDPVQRKTYSLELPELDGCRV-CYTKDG-WLLLNRQDWRRLDGNHIFSLFNPFTRDLITL 70
+DP+Q K Y+ L +D ++ C G W+L+ +D F L N FTR+ I L
Sbjct: 62 FDPIQDKIYTKNLLGIDLSKIHCLASYGNWILI-------VDPRLDFYLLNVFTRETINL 114
Query: 71 PKF------DRTYQ-------------------IAAFSCAPTSTGCVILIFRRVGSSLV- 104
P DR ++ + ++ ++ + R G +V
Sbjct: 115 PSLESSLRGDRPFRFIRNDDESGYFLELYGTKFLLTWNDFRFEKTAILWVNGRNGDYVVA 174
Query: 105 -AISTCY----------PGEKEWTTVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFD 153
AI Y +K W+ ++C+ C+ + + + Y + ++ + D
Sbjct: 175 WAIKQFYIFSYKKIGNDDDDKRWS-------ITCTQCEDMAYKDNKLYVYTFDHYINILD 227
Query: 154 PLERTWTVFKVPPPKCLAESSTAKNWSKGKFMIEHKGNIFVVHICCGEDP--IIFKLDLT 211
+ P S K + + +G + +V G D I+KL+L
Sbjct: 228 FSGNSPKEPMEENPYLSHPFSFVDAIYKLRLAVTREGKVLIVLSLVGLDKKICIYKLNLK 287
Query: 212 LMEWKEVRSL 221
+ +W+ V SL
Sbjct: 288 IGDWEIVESL 297
>At2g43440 hypothetical protein
Length = 779
Score = 32.0 bits (71), Expect = 0.42
Identities = 26/111 (23%), Positives = 43/111 (38%), Gaps = 10/111 (9%)
Query: 110 YPGEKEWTTVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKC 169
+ G++E ++ DA CD L+ C+++ GW V +P F PP
Sbjct: 83 FEGDEEIVYLHCDAAQPSMTCDGLL-------CITEPGWFNVLNPSAGQLRRF---PPGP 132
Query: 170 LAESSTAKNWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRS 220
+NW G G +V +C + LD+ W ++RS
Sbjct: 133 GPVKGPQENWLLGFGRDNVTGRYKIVRMCFHDCYEFGILDIETGVWSKLRS 183
>At1g44080 hypothetical protein
Length = 311
Score = 32.0 bits (71), Expect = 0.42
Identities = 25/85 (29%), Positives = 39/85 (45%), Gaps = 18/85 (21%)
Query: 181 KGKFMIEHKGNIFVVHICCGEDPI------------IFKLDLTLMEWKEVRSLNGVTLFA 228
K K ++E+ G + VVH + + ++K+D L+EW EV SL L
Sbjct: 188 KEKRLVEYCGELCVVHRFYKKFCVKRVLTERTVCFKVYKMDKNLVEWVEVSSLGDKALIV 247
Query: 229 S----FLSSHSRTYATGIMRNSVYF 249
+ FL S Y G + N++YF
Sbjct: 248 ATDNCFLVLASEYY--GCLENAIYF 270
>At1g27420 hypothetical protein
Length = 346
Score = 32.0 bits (71), Expect = 0.42
Identities = 30/117 (25%), Positives = 49/117 (41%), Gaps = 23/117 (19%)
Query: 99 VGSSLVAISTCYPGEKEWTTVYYDAELSCSMCDKLVFSNGLFYCLSDR---GWLGVFDPL 155
VG+ I P + W + ++E S S+ V N +++ DR G LGVFDP
Sbjct: 220 VGNGSRFIDIYDPKTQTWEEL--NSEQSVSVYSYTVVRNKVYFM--DRNMPGRLGVFDPE 275
Query: 156 ERTWTVFKVPPPKCLAESSTAKNWSKGKFMIE---HKGNIFVVHICCGEDPIIFKLD 209
E +W+ VPP +G F + + + CG + +++ LD
Sbjct: 276 ENSWSSVFVPP-------------REGGFWVRLGVWNNKVLLFSRVCGHETLMYDLD 319
>At3g03730 hypothetical protein
Length = 393
Score = 31.6 bits (70), Expect = 0.54
Identities = 33/164 (20%), Positives = 60/164 (36%), Gaps = 9/164 (5%)
Query: 130 CDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAKNWSKGKFMIEHK 189
C +VF G Y L+ + VFD V PP S ++ +
Sbjct: 222 CINMVFKEGKLYVLNPARNISVFDFSGGHSPVEYATPP------SPNDDYYVRNLAVTLS 275
Query: 190 GNIFVVHICCGEDPI-IFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRTYATGIMRNSVY 248
G + ++ + + ++K+D EW+ ++S+ L + G+MRN +Y
Sbjct: 276 GEVLIISSNPKKCFVKLYKIDPKSSEWRLIKSIGDEALILDL--GITVAAKDGVMRNCIY 333
Query: 249 FPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPP 292
F + +S DD+ + K + FE+ P
Sbjct: 334 FSHHELLRYKGVSLCNDDKYGICIYHIKTKKKVQEFEHLTTSSP 377
>At3g24610 hypothetical protein
Length = 445
Score = 30.8 bits (68), Expect = 0.93
Identities = 26/107 (24%), Positives = 40/107 (37%), Gaps = 18/107 (16%)
Query: 148 WLGVFDPLERTWTVFKVPPPKCLAESSTAKNWSKGKF------MIEHKGN---------- 191
W VFDP +TW +P P + +W G + I KGN
Sbjct: 223 WGEVFDPKTQTWADMSIPKPVREEKIYVVDSWDVGSYYYLPSKSIWEKGNQDSKRSKDWC 282
Query: 192 -IFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFA-SFLSSHSR 236
I + CG D I+ + ++W + ++ +F FLS R
Sbjct: 283 LIDKLIYSCGNDGGIYWCEAGELDWCDAVGIDWREVFGLEFLSKELR 329
>At5g09580 putative protein
Length = 393
Score = 30.4 bits (67), Expect = 1.2
Identities = 26/88 (29%), Positives = 37/88 (41%), Gaps = 7/88 (7%)
Query: 57 FSLFNPFTRDLITLPKFDRTYQI-----AAFSCAPTSTGCVILIFRRVGSSLVAISTCYP 111
FS PF + + L F+R Y + F+C T+ C I+ F S+L I
Sbjct: 108 FSSLIPFRVEDLCLEGFERCYLLDFVVPKDFACQKTA--CEIVCFDHRNSALKRIGLIKE 165
Query: 112 GEKEWTTVYYDAELSCSMCDKLVFSNGL 139
K+ +Y D E S S FS+ L
Sbjct: 166 EHKKRLKIYVDTETSSSKAVYKYFSSKL 193
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,927,601
Number of Sequences: 26719
Number of extensions: 347775
Number of successful extensions: 936
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 876
Number of HSP's gapped (non-prelim): 39
length of query: 299
length of database: 11,318,596
effective HSP length: 99
effective length of query: 200
effective length of database: 8,673,415
effective search space: 1734683000
effective search space used: 1734683000
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139854.4


