

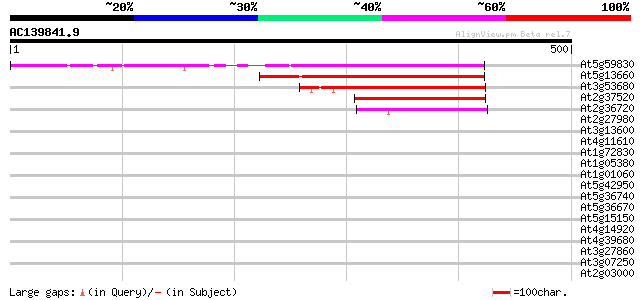
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.9 + phase: 0
(500 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59830 unknown protein 309 3e-84
At5g13660 unknown protein 183 2e-46
At3g53680 putative protein 142 6e-34
At2g37520 unknown protein 130 2e-30
At2g36720 putative PHD-type zinc finger protein 75 6e-14
At2g27980 hypothetical protein 41 0.001
At3g13600 hypothetical protein 37 0.033
At4g11610 phosphoribosylanthranilate transferase like protein 36 0.043
At1g72830 unknown protein 34 0.21
At1g05380 unknown protein 33 0.37
At1g01060 DNA-binding protein, putative 32 0.62
At5g42950 putative protein 32 0.82
At5g36740 putative protein 32 0.82
At5g36670 putative protein 32 0.82
At5g15150 homeobox protein 32 0.82
At4g14920 hypothetical protein 32 0.82
At4g39680 unknown protein 31 1.4
At3g27860 hypothetical protein 30 3.1
At3g07250 putative RNA-binding protein 30 3.1
At2g03000 hypothetical protein 30 3.1
>At5g59830 unknown protein
Length = 425
Score = 309 bits (791), Expect = 3e-84
Identities = 185/428 (43%), Positives = 249/428 (57%), Gaps = 38/428 (8%)
Query: 1 MVKGSGHVSDREPAFDNPSKIEPKRPHQWLIDATEGDFLPNKKQAIEDANERSSSGFSNV 60
++K + H S+ + +D+ ++ + KRPH W +D++ + PNKKQA++D G SNV
Sbjct: 10 VMKNNEHTSEEDSVYDHSTRDDSKRPHPWFVDSSRSEMFPNKKQAVQDPVV--GLGKSNV 67
Query: 61 NFTPWENNHNFHSDPSHQNQLIDRLFGSET--RPVNFTEKNTYVSGDGSDVRSKMIANHY 118
WE++ F S NQ +DRL G+E RP+ F +++ G ++K IA Y
Sbjct: 68 GLPLWESSSVFQSV---SNQFMDRLLGAEMPPRPLLFGDRDR-TEGCSHHHQNKSIAESY 123
Query: 119 GDGASFGLSISHSTEDSEPCMNFGGIKKVKVNQVNP--SDVQAPEQHNFDRQSTGDLHHV 176
+ S LSIS+ E + C G +K+ V++V S A E H+ + + +
Sbjct: 124 MEDTSVELSISNGVEVAGGCFGGDGNRKLPVSRVKETMSTHVALEGHSQRKIESSSIQAC 183
Query: 177 YHGEVETRSGSIGLAYGSGDARIRPFGTPYGKVDNTVLSIAESYNKDETNIISFGGFPDE 236
E S I A G PYG D+ I+FG DE
Sbjct: 184 SR---ENESSYINFALA---------GHPYGNEDSQG--------------ITFGEINDE 217
Query: 237 RGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASN-SDVVASTPLVTTKKPESVSKNK 295
GV S +Y Q Y Q + +++E +S S V S V S+ K K
Sbjct: 218 HGVGSTSNVVGNY-QSYVQDPIGTLDIVYDQETGSSQTSSGVVSEQQVAKPSLGSLPKTK 276
Query: 296 QDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREELRGIIKGTTYLCGCQSCNY 355
+ KS++KE+ +FP+NVRSLISTGMLDGVPVKYVSV+REELRG+IKG+ YLCGCQ+C++
Sbjct: 277 AEAKSSKKEASTSFPSNVRSLISTGMLDGVPVKYVSVSREELRGVIKGSGYLCGCQTCDF 336
Query: 356 AKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPIN 415
K LNA+ FE+HAGCK+KHPNNHIYFENGKTIYQIVQELR+TPES LFD IQT+FG+PIN
Sbjct: 337 TKVLNAYAFERHAGCKTKHPNNHIYFENGKTIYQIVQELRNTPESILFDVIQTVFGSPIN 396
Query: 416 QKAFRIWK 423
QKAFRIWK
Sbjct: 397 QKAFRIWK 404
>At5g13660 unknown protein
Length = 536
Score = 183 bits (464), Expect = 2e-46
Identities = 94/202 (46%), Positives = 128/202 (62%), Gaps = 3/202 (1%)
Query: 223 DETNIISFGGFPDERGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASNSDVVASTPL 282
D + +SFG E + S R + +YE + ++ + E E + S + P
Sbjct: 314 DRSETLSFGDCQKETAMGSSVRVSNNYENFSHDPAI--TKDPLHIEAEENMSFECRNPPY 371
Query: 283 VTTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREE-LRGII 341
+ + + +D K+ +K S NTFP+NV+SL+STG+ DGV VKY S +RE L+G+I
Sbjct: 372 ASPRVDTLLVPKIKDTKTAKKGSTNTFPSNVKSLLSTGIFDGVTVKYYSWSRERNLKGMI 431
Query: 342 KGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESS 401
KGT YLCGC +C K LNA+EFE+HA CK+KHPNNHIYFENGKTIY +VQEL++TP+
Sbjct: 432 KGTGYLCGCGNCKLNKVLNAYEFEQHANCKTKHPNNHIYFENGKTIYGVVQELKNTPQEK 491
Query: 402 LFDTIQTIFGAPINQKAFRIWK 423
LFD IQ + G+ IN K F WK
Sbjct: 492 LFDAIQNVTGSDINHKNFNTWK 513
Score = 34.7 bits (78), Expect = 0.13
Identities = 50/241 (20%), Positives = 89/241 (36%), Gaps = 48/241 (19%)
Query: 12 EPAFDNPSKIEPKRPH-QWLIDATEGDFLPNKKQAIEDANERSSSGFSNVNFTPWENNHN 70
E + S++E KR H QWL + + + NK+Q + + + +++N +PW+ +
Sbjct: 14 EIPYSGSSRMELKRSHHQWLTEESSSELFSNKRQQVVEID-------AHMNLSPWDTS-- 64
Query: 71 FHSDPSHQNQLIDRLFGSETRPVNFTEKNTYVSGDGSDVRSKMIANHYGDGASFGLSISH 130
PSH D LF +++ +G + + +SFGL ++H
Sbjct: 65 --LVPSH---FTDCLFDDPA-----IAHTSHLLRNGRNYTEEQC----NPVSSFGLPLAH 110
Query: 131 STEDSEPCMNFGGIKKVK---------VNQVNPSDVQAPEQHNFDRQST----------G 171
N I KV Q + + N ++ST
Sbjct: 111 HGSS----FNLDTINKVSNVPEFMVQLYGQGISTSFETAPSFNSGQESTTLSFGQTFSNT 166
Query: 172 DLHHVYHGEVETRS-GSIGLAYGSGDARIRPFGTPYGKVDNTVLSIAESYNKDETNIISF 230
D + G+ +++ G+ + + + P G Y K D VLS K N +S
Sbjct: 167 DRSFILPGQFASKTDGNFIRNFNNEGVGVVPIGDYYDKGDENVLSTFHPLEKGVENFLSM 226
Query: 231 G 231
G
Sbjct: 227 G 227
>At3g53680 putative protein
Length = 839
Score = 142 bits (357), Expect = 6e-34
Identities = 72/174 (41%), Positives = 107/174 (61%), Gaps = 9/174 (5%)
Query: 259 HVSTTAHEK---ELEASNSDVVASTPLVTTKK----PESVSKNKQDIKSTRKESPNTFPT 311
H+STT K +L N D P++ P +V+ + +K +K F +
Sbjct: 126 HLSTTGITKITFKLSKRNEDF-CDLPMIQEHTWEGYPSNVASSTLGVKMLKKIDSTNFLS 184
Query: 312 NVRSLISTGMLDGVPVKYVSV-AREELRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGC 370
NV+ L+ TG+LDG VKY+S A EL+GII YLCGC +C+++K L A+EFE+HAG
Sbjct: 185 NVKKLLGTGILDGARVKYLSTSAARELQGIIHSGGYLCGCTACDFSKVLGAYEFERHAGG 244
Query: 371 KSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWKG 424
K+KHPNNHIY ENG+ +Y ++QELR P L + I+ + G+ ++++ F+ WKG
Sbjct: 245 KTKHPNNHIYLENGRPVYNVIQELRIAPPDVLEEVIRKVAGSALSEEGFQAWKG 298
Score = 33.9 bits (76), Expect = 0.21
Identities = 20/79 (25%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query: 317 ISTGMLDGVPVKYVSVAREELRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKS-KHP 375
+ G+ DG + Y +++ L+G +G+ +C C C+ ++ +FE HAG + P
Sbjct: 399 LPNGLPDGTELAYYVKSQKLLQGYKQGSGIVCSC--CD--TKISPSQFEAHAGMAGRRQP 454
Query: 376 NNHIYFENGKTIYQIVQEL 394
I+ +G +++ I L
Sbjct: 455 YRRIHISSGLSLHDIAVSL 473
>At2g37520 unknown protein
Length = 825
Score = 130 bits (327), Expect = 2e-30
Identities = 56/118 (47%), Positives = 85/118 (71%), Gaps = 1/118 (0%)
Query: 308 TFPTNVRSLISTGMLDGVPVKYVSVAR-EELRGIIKGTTYLCGCQSCNYAKGLNAFEFEK 366
++P+NV+ L+ TG+L+G VKY+S +L GII YLCGC +CN++K L+A+EFE+
Sbjct: 162 SYPSNVKKLLETGILEGARVKYISTPPVRQLLGIIHSGGYLCGCTTCNFSKVLSAYEFEQ 221
Query: 367 HAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWKG 424
HAG K++HPNNHI+ EN + +Y IVQEL++ P L + I+ + G+ +N++ R WKG
Sbjct: 222 HAGAKTRHPNNHIFLENRRAVYNIVQELKTAPRVVLEEVIRNVAGSALNEEGLRAWKG 279
Score = 29.6 bits (65), Expect = 4.0
Identities = 15/40 (37%), Positives = 23/40 (57%)
Query: 32 DATEGDFLPNKKQAIEDANERSSSGFSNVNFTPWENNHNF 71
+ +GD P+KKQA E +N+ +S SN +P E+ F
Sbjct: 20 EGNKGDHFPSKKQAKEASNDDITSEISNPVASPVESTSLF 59
>At2g36720 putative PHD-type zinc finger protein
Length = 958
Score = 75.5 bits (184), Expect = 6e-14
Identities = 42/120 (35%), Positives = 65/120 (54%), Gaps = 3/120 (2%)
Query: 310 PTNVRSLISTGMLDGVPVKYVSVAREE---LRGIIKGTTYLCGCQSCNYAKGLNAFEFEK 366
P VR L TG+LDG+ V Y+ + + LRGII+ LC C SC++A ++ +FE
Sbjct: 261 PETVRDLFETGLLDGLSVVYMGTVKSQAFPLRGIIRDGGILCSCSSCDWANVISTSKFEI 320
Query: 367 HAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWKGKE 426
HA + + + +I FENGK++ ++ R+TP +L TI +K F + KE
Sbjct: 321 HACKQYRRASQYICFENGKSLLDVLNISRNTPLHALEATILDAVDYASKEKRFTCKRCKE 380
>At2g27980 hypothetical protein
Length = 1008
Score = 41.2 bits (95), Expect = 0.001
Identities = 32/133 (24%), Positives = 59/133 (44%), Gaps = 27/133 (20%)
Query: 286 KKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREELRGIIKGTT 345
+ P + NK + K+ FP ++ + G+L+G+ V YV RG
Sbjct: 357 QSPSVTTPNK---RGRPKKFLRNFPAKLKDIFDCGILEGLIVYYV-------RG------ 400
Query: 346 YLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDT 405
AK ++ FE HA +K P +I E+G T+ ++ + P ++L +
Sbjct: 401 ----------AKVVSPAMFELHASSNNKRPPEYILLESGFTLRDVMNACKENPLATLEEK 450
Query: 406 IQTIFGAPINQKA 418
++ + G PI +K+
Sbjct: 451 LRVVVG-PILKKS 462
Score = 31.2 bits (69), Expect = 1.4
Identities = 13/28 (46%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query: 364 FEKHAGCKSKH-PNNHIYFENGKTIYQI 390
FE HAGC S+ P HIY NG +++++
Sbjct: 603 FEAHAGCASRRKPFQHIYTTNGVSLHEL 630
>At3g13600 hypothetical protein
Length = 605
Score = 36.6 bits (83), Expect = 0.033
Identities = 31/107 (28%), Positives = 52/107 (47%), Gaps = 10/107 (9%)
Query: 220 YNKDETNIISFGGFPDERGV--ISVGRAATDYEQ--LYNQSSVHVSTTAHEKELEASNS- 274
Y E N + F F E V V + TD ++ +Y Q S H+ + E++LEA +
Sbjct: 346 YQPTEENFMDFLSFLRENDVDITDVKMSPTDEDEFSIYKQRSTHMRNHSLEEDLEAEKTI 405
Query: 275 ----DVVASTPLVTTKKPESVSKNKQDIKSTRK-ESPNTFPTNVRSL 316
V S T + ES+S+ + D+++ K ES +TF ++S+
Sbjct: 406 SFQDKVDPSGEEQTLMRNESISRKQSDLETPEKMESFSTFGDEIQSV 452
>At4g11610 phosphoribosylanthranilate transferase like protein
Length = 1011
Score = 36.2 bits (82), Expect = 0.043
Identities = 35/156 (22%), Positives = 58/156 (36%), Gaps = 19/156 (12%)
Query: 16 DNPSKIEPKRPHQWLIDATEGDF-----LPNKKQAIEDANERSSSGFSNVNFTPWENNHN 70
D+P ++P P ++ LPN Q E ++ + + ++NHN
Sbjct: 145 DHPDNLDPALPRAMNVEHRSDKRHVFYNLPNSAQ--EHQHQHPQGPNQSSSLAAEQDNHN 202
Query: 71 FHSD---PSHQNQLIDRLFGSETRPVNFTEKNTYVSGDGSDVRSKMIANHYGDGASFGLS 127
H P HQ +D + RP ++ S +D K + H G G G
Sbjct: 203 EHHHHYVPKHQ---VDEMRSEPARPSKLVHAHSIASAQPADFALKETSPHLGGGRVVGGR 259
Query: 128 ISH------STEDSEPCMNFGGIKKVKVNQVNPSDV 157
+ H ST D M F ++ VK ++ D+
Sbjct: 260 VIHKDKTATSTYDLVERMYFLYVRVVKARELPIMDI 295
>At1g72830 unknown protein
Length = 340
Score = 33.9 bits (76), Expect = 0.21
Identities = 19/67 (28%), Positives = 30/67 (44%), Gaps = 7/67 (10%)
Query: 16 DNPSKIEPKRPHQWLIDATEGDFLPNKKQAIEDANERSSSGFSNVNFTPWENNHNFHSDP 75
DNPS+ Q A G + +K + + S +GF N++F P + N +FH
Sbjct: 87 DNPSR-------QISFSAKSGSEITQRKGFASNPKQGSMTGFPNIHFAPAQANFSFHYAD 139
Query: 76 SHQNQLI 82
H L+
Sbjct: 140 PHYGGLL 146
>At1g05380 unknown protein
Length = 1138
Score = 33.1 bits (74), Expect = 0.37
Identities = 22/57 (38%), Positives = 27/57 (46%), Gaps = 4/57 (7%)
Query: 333 AREELRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQ 389
A+ L G I C C S K L FE HAG KS P +IY E+G ++ Q
Sbjct: 544 AKVMLEGWITREGIHCDCCS----KILTVSRFEIHAGSKSCQPFQNIYLESGASLLQ 596
>At1g01060 DNA-binding protein, putative
Length = 645
Score = 32.3 bits (72), Expect = 0.62
Identities = 20/70 (28%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Query: 250 EQLYNQSSVHVSTTAHEKELEASNSDVVASTPL---VTTKKPESVSKNKQDIKSTRKESP 306
E++ ++VH S TA +K L S ++TP T + + K+K+D+K T + P
Sbjct: 444 EEVVVTAAVHDSNTAQKKNL-VDRSSCGSNTPSGSDAETDALDKMEKDKEDVKETDENQP 502
Query: 307 NTFPTNVRSL 316
+ N R +
Sbjct: 503 DVIELNNRKI 512
>At5g42950 putative protein
Length = 1714
Score = 32.0 bits (71), Expect = 0.82
Identities = 33/135 (24%), Positives = 54/135 (39%), Gaps = 11/135 (8%)
Query: 212 TVLSIAESYNKDETNIISFGGFPDERGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEA 271
T I ES +E + + G +G +S+ + + V V+ EK+
Sbjct: 1055 TAPEIVESKLLEEQSKDMYAG----KGEVSIELSGETPATEVKNNDVSVARKTSEKKSRK 1110
Query: 272 SNS----DVVAST---PLVTTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDG 324
+ D+ ST PL TKKP+ S + +IK K+S +T N LI +
Sbjct: 1111 QRAKQAADLAKSTSRAPLQETKKPQPGSADDSEIKGKTKKSADTLIDNDTHLIKSSTATA 1170
Query: 325 VPVKYVSVAREELRG 339
+S + +RG
Sbjct: 1171 SNTSQMSSEVDSVRG 1185
>At5g36740 putative protein
Length = 960
Score = 32.0 bits (71), Expect = 0.82
Identities = 21/67 (31%), Positives = 29/67 (42%), Gaps = 5/67 (7%)
Query: 337 LRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQE-LR 395
L GII C C C+ + +FE HAG P +Y E G ++ Q + E +
Sbjct: 573 LEGIITKEGIRCNC--CDEV--FSVLDFEVHAGGNRNQPFKSLYLEGGNSLLQCLHESMN 628
Query: 396 STPESSL 402
ES L
Sbjct: 629 KQSESQL 635
>At5g36670 putative protein
Length = 1030
Score = 32.0 bits (71), Expect = 0.82
Identities = 21/67 (31%), Positives = 29/67 (42%), Gaps = 5/67 (7%)
Query: 337 LRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQE-LR 395
L GII C C C+ + +FE HAG P +Y E G ++ Q + E +
Sbjct: 420 LEGIITKEGIRCNC--CDEV--FSVLDFEVHAGGNRNQPFKSLYLEGGNSLLQCLHESMN 475
Query: 396 STPESSL 402
ES L
Sbjct: 476 KQSESQL 482
>At5g15150 homeobox protein
Length = 293
Score = 32.0 bits (71), Expect = 0.82
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 7/88 (7%)
Query: 16 DNPSKI-EPKRPHQWLIDATEGDFLPNKKQAIEDANERSSSGFSNVNFTPWENNHNFHSD 74
DN S + K+ H L+ + D +K++ + E + + +SN T ENNHN +S
Sbjct: 168 DNDSLLAHNKKLHAELVALKKHD----RKESAKIKREFAEASWSNNGST--ENNHNNNSS 221
Query: 75 PSHQNQLIDRLFGSETRPVNFTEKNTYV 102
++ +I LF S R T +T++
Sbjct: 222 DANHVSMIKDLFPSSIRSATATTTSTHI 249
>At4g14920 hypothetical protein
Length = 1040
Score = 32.0 bits (71), Expect = 0.82
Identities = 27/109 (24%), Positives = 46/109 (41%), Gaps = 12/109 (11%)
Query: 284 TTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDG------VPVKYVSVAREE- 336
++K S ++ ++ ES P++ + + ++D V Y++ R
Sbjct: 514 SSKHGRSTLLVRRSVRGDNSESDGFVPSSEKRTVLAWLIDSGTLQLSEKVMYMNQRRTRA 573
Query: 337 -LRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENG 384
L G I CGC S K L +FE HAG K + P +I+ +G
Sbjct: 574 MLEGWITRDGIHCGCCS----KILAVSKFEIHAGSKLRQPFQNIFLNSG 618
>At4g39680 unknown protein
Length = 633
Score = 31.2 bits (69), Expect = 1.4
Identities = 22/99 (22%), Positives = 40/99 (40%), Gaps = 1/99 (1%)
Query: 210 DNTVLSIAESYNKDETNIISFGGFPDERGVISVGRAATDYEQLYNQSSVHVSTTAHEKEL 269
D V+ +A S +K E N F G A + +YNQ S + T E +
Sbjct: 169 DAAVVQVASSEHKSENNE-PFSGLDGGDSKAQPSEAVLEKSAMYNQVSEVIPVTGFEVKS 227
Query: 270 EASNSDVVASTPLVTTKKPESVSKNKQDIKSTRKESPNT 308
+ ++D V++ + K + K + + + P+T
Sbjct: 228 DCISTDSVSNNEKIELKDNKIADNVKLEQNVNKFQEPST 266
>At3g27860 hypothetical protein
Length = 652
Score = 30.0 bits (66), Expect = 3.1
Identities = 23/93 (24%), Positives = 39/93 (41%), Gaps = 14/93 (15%)
Query: 39 LPNKKQAIEDANERSSSGFSNVNFTPWENNHNFHSDPSHQNQLIDRLFGSETRPVNFTEK 98
+ N K+ E + SG S++ E + FH D ++G E +
Sbjct: 29 MENVKEDSEQIGDNVRSGVSSLRDNFEELENGFHVG--------DFVWGEEANSQQWWPG 80
Query: 99 NTYVSGDGSDV------RSKMIANHYGDGASFG 125
Y S D SD+ + K++ ++GDG+ FG
Sbjct: 81 QIYDSLDASDLALKTMQKGKLLVAYFGDGSFFG 113
>At3g07250 putative RNA-binding protein
Length = 946
Score = 30.0 bits (66), Expect = 3.1
Identities = 40/163 (24%), Positives = 58/163 (35%), Gaps = 30/163 (18%)
Query: 96 TEKNTYVSGDGSDVRSKMIANHYGDG--------ASFGLSISHSTEDSEPCMNF---GGI 144
TE N +GD D+ S + N YG G + G I S+ S+P + GG
Sbjct: 88 TEYNGVSNGDDMDLSSINMCNGYGSGRLKESSKDVTIGSPICTSSSVSDPSSSSRQEGGS 147
Query: 145 KKVKVNQVNPSDVQAP--------EQ--HNFDRQSTGDLHHVYHGEVETRSGSIGLAY-- 192
+ V++P EQ HN D + + V SG +
Sbjct: 148 GDIGAQSGQTKQVRSPVNNILTGTEQTVHNTDGSKDAETQEWSNSSVTKESGKPPKPHTN 207
Query: 193 GSGDARIRPF-------GTPYGKVDNTVLSIAESYNKDETNII 228
+G+ + PF T G TV Y+K E +II
Sbjct: 208 SNGNGSLLPFAQMPCVTSTGNGPEGKTVNGFLYRYSKSEISII 250
>At2g03000 hypothetical protein
Length = 535
Score = 30.0 bits (66), Expect = 3.1
Identities = 31/127 (24%), Positives = 57/127 (44%), Gaps = 5/127 (3%)
Query: 225 TNIISFGGFPDERGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASNSDVVASTPLV- 283
T++ S G FPD+R + ++ATD N+S + + SNS V T +
Sbjct: 50 TSMSSGGDFPDDRFFHARHQSATD-RAASNRSRAIMPISTTNVPSSTSNSPRVLQTSMSR 108
Query: 284 --TTKKPESVSKNKQDIKSTRKESPNTFPT-NVRSLISTGMLDGVPVKYVSVAREELRGI 340
T+ S ++ Q S R+ P++ T ++++ ST + + V + EE+
Sbjct: 109 RGTSPMSSSTRRSVQASMSARETVPSSTSTRSMQTSTSTPEIMPTSSRNVITSSEEVANT 168
Query: 341 IKGTTYL 347
T+Y+
Sbjct: 169 FTQTSYI 175
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,269,549
Number of Sequences: 26719
Number of extensions: 563737
Number of successful extensions: 1190
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 1161
Number of HSP's gapped (non-prelim): 43
length of query: 500
length of database: 11,318,596
effective HSP length: 103
effective length of query: 397
effective length of database: 8,566,539
effective search space: 3400915983
effective search space used: 3400915983
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139841.9


