

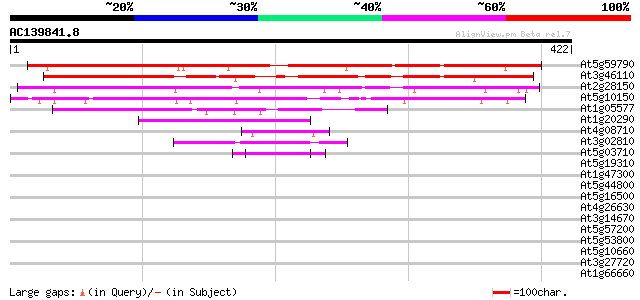
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.8 + phase: 0
(422 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59790 unknown protein 391 e-109
At3g46110 unknown protein 298 4e-81
At2g28150 unknown protein 256 1e-68
At5g10150 unknown protein 160 9e-40
At1g05577 hypothetical protein 115 6e-26
At1g20290 hypothetical protein 49 4e-06
At4g08710 putative protein 45 6e-05
At3g02810 putative protein kinase 44 1e-04
At5g03710 putative protein 43 3e-04
At5g19310 homeotic gene regulator - like protein 41 0.001
At1g47300 hypothetical protein 41 0.001
At5g44800 helicase-like protein 40 0.002
At5g16500 protein kinase-like protein 40 0.002
At4g26630 unknown protein 40 0.002
At3g14670 hypothetical protein 40 0.002
At5g57200 putative protein 40 0.003
At5g53800 unknown protein 39 0.004
At5g10660 vacuolar calcium binding protein - like 39 0.004
At3g27720 hypothetical protein 39 0.004
At1g66660 hypothetical protein 39 0.004
>At5g59790 unknown protein
Length = 423
Score = 391 bits (1004), Expect = e-109
Identities = 226/414 (54%), Positives = 274/414 (65%), Gaps = 43/414 (10%)
Query: 14 RETSPERTKVWTEP--KPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIR 71
++TSP+R ++W+EP KP RKV VVYYL RNG L+HPHF+EV LSS GLYLKDVI R
Sbjct: 24 QDTSPDRNRIWSEPRLKPVVNRKVPVVYYLCRNGQLDHPHFIEVTLSSHDGLYLKDVINR 83
Query: 72 LNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEH------ 125
LN LRGKGM ++YSWSSKRSYKNGFVWHDL+E DFI+P QG +Y+LKGSE+L+
Sbjct: 84 LNDLRGKGMASLYSWSSKRSYKNGFVWHDLSEDDFIFPVQGQEYVLKGSEVLDSCLISNP 143
Query: 126 ----ETTA-------RPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYK----SEPLGEN 170
ET++ P D P VI RRRNQSWSSIDL+EY+VYK S +
Sbjct: 144 RSLLETSSFRDPRSLNPDKNSGDDIPAVINRRRNQSWSSIDLSEYKVYKATESSAESTQR 203
Query: 171 IAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPP 230
+AADASTQT+++RRRRR +EE EE + +NQSTELSR+EISPP
Sbjct: 204 LAADASTQTDDRRRRRRPAKEEIEEV-------------KSPASYENQSTELSRDEISPP 250
Query: 231 PSDSSPETLGTLMKVDGRLGLR--SVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKE 288
PSDSSPETL L+K DGRL LR ++ TVES SGRMRAS+VL+QL+SCG +SFKE
Sbjct: 251 PSDSSPETLENLIKADGRLILRPSESSTDHRTVESLSSGRMRASAVLMQLISCGTMSFKE 310
Query: 289 GSGASSGKDQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLIE 348
G KDQ +L G + RG ++ + V E+ RV+LEDKEYFSGSLIE
Sbjct: 311 -CGPVLLKDQGLALNGRSGCTITRGAEDNGEERVDK--ELKSFGRVQLEDKEYFSGSLIE 367
Query: 349 TKKVEAPAFKRSSSYNADSSSRL--QIMEHEGDMVRAKCIPRKSKTLLTKKEEG 400
TKK PA KRSSSYNAD SR+ + E + VRAKCIPRK K + + G
Sbjct: 368 TKKELVPALKRSSSYNADRCSRMGPTTEKDEEEAVRAKCIPRKPKPVAKRNNGG 421
>At3g46110 unknown protein
Length = 343
Score = 298 bits (763), Expect = 4e-81
Identities = 187/379 (49%), Positives = 239/379 (62%), Gaps = 59/379 (15%)
Query: 26 EPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYS 85
+ KP R V VVYYLSRNG L+HPHF+EVPLSS +GLYLKDVI RLN LRG GM +YS
Sbjct: 11 DSKPSRERIVPVVYYLSRNGRLDHPHFIEVPLSSHNGLYLKDVINRLNDLRGNGMACLYS 70
Query: 86 WSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITR 145
WSSKR+YKNGFVW+DL++ DFI+P G +Y+LKGS+IL+ L+ S + +T
Sbjct: 71 WSSKRTYKNGFVWYDLSDEDFIFPVHGQEYVLKGSQILD--------LDNNSGNFSAVTH 122
Query: 146 RRNQSWSSIDLNEYRVYKSEPLG----ENIAADASTQTEEKRRRRRVLREEDEEQEEEEK 201
RRNQSWSS+D Y+VYK+ L ++ DASTQT+++RRR K
Sbjct: 123 RRNQSWSSVD--HYKVYKASELNAEATRKLSMDASTQTDDRRRR---------------K 165
Query: 202 EKNDEIENEGETENQNQSTELSREEI-SPPPSDSSPETLGTLMKVDGRLGLRSVPKE-NP 259
DE+ N+ TELSREEI SPP SDSSPETL +LM+ DGRL L +E N
Sbjct: 166 SPVDEV---------NEVTELSREEITSPPQSDSSPETLESLMRADGRLILLQEDQELNR 216
Query: 260 TVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASSGKDQRFSLVGHYKSRLPRGGGNHVG 319
TVE +MR S+VL+QL+SCGA+SFK K + G+ +S RG GN+
Sbjct: 217 TVE-----KMRPSAVLMQLISCGAMSFK--------KCGPTLMNGNTRSTAVRGTGNYRL 263
Query: 320 KEVGTLVEIPDLNRVRLEDKEYFSGSLIE--TKKVEAPAFKRSSSYNADSSSRLQI-MEH 376
+ E+ RV+LE+KEYFSGSLI+ +KK PA KRSSSYN D SSR+ + E
Sbjct: 264 ERAEK--ELKSFGRVKLEEKEYFSGSLIDESSKKELVPALKRSSSYNIDRSSRMGLTKEK 321
Query: 377 EG-DMVRAKCIPRKSKTLL 394
EG ++ RA IPR +++
Sbjct: 322 EGEELARANFIPRNPNSVV 340
>At2g28150 unknown protein
Length = 540
Score = 256 bits (655), Expect = 1e-68
Identities = 180/481 (37%), Positives = 250/481 (51%), Gaps = 105/481 (21%)
Query: 7 SRGRPGTRETSPERTKVWTEPKPKTP---RKVSVVYYLSRNGHLEHPHFMEVPLSSPHGL 63
+R + +RE SPER KVWTE PK +KV +VYYLS+N LEHPHFMEV +SSP+GL
Sbjct: 3 ARMKKYSREVSPERAKVWTEKSPKYHQKIKKVQIVYYLSKNRQLEHPHFMEVLISSPNGL 62
Query: 64 YLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEIL 123
YL+DVI RLN+LRG+GM +MYSWSSKRSY+NGFVWHDL+E D I P G++Y+LKGSE+
Sbjct: 63 YLRDVIERLNVLRGRGMASMYSWSSKRSYRNGFVWHDLSEDDLILPANGNEYVLKGSELF 122
Query: 124 ----------------------------------------------------EHETTARP 131
E + + P
Sbjct: 123 DESNSDHFSPIVNLATQNMKQIVVEPPSSRSMDDSSSSSSMNNGKGTNKHSHEDDELSPP 182
Query: 132 PLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRR----- 186
L S S V R ++ SS L EY+VYKSE L ADASTQT+E R
Sbjct: 183 ALRSVSSSGVSPDSRDAKNSSSWCLAEYKVYKSEGL-----ADASTQTDETVSGRSKTPI 237
Query: 187 ----RVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSS------P 236
R + +++ E E +N+ + + +S E+SR +SPP S+S+
Sbjct: 238 ETFSRGVSTDEDVSSEPETSENNLVSEASCAGKERESAEISRNSVSPPFSNSASSLGGKT 297
Query: 237 ETLGTLMKVD--GRLGLRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASS 294
+TL +L++ D R + +E+ + + P R+RAS++L+QL+SCG++S
Sbjct: 298 DTLESLIRADVSKMNSFRILEQEDVRMPAIP--RLRASNMLMQLISCGSISV-------- 347
Query: 295 GKDQRFSLV-------GHYKSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLI 347
KD F LV GH K P + + ++ L E P L +R+E+KEYFSGSL+
Sbjct: 348 -KDNNFGLVPTYKPKFGHSKFPSPFFSSSFMMGDLDRLSETPSLMGLRMEEKEYFSGSLV 406
Query: 348 ETKKVEAPA------FKRSSSYNADSSSRLQIMEHEGDMV--RAKCIP--RKSKTLLTKK 397
ETK + A KRSSSYN D +S + GD +K P RK+ ++L K+
Sbjct: 407 ETKLQKKDAADSNASLKRSSSYNGDRASNQMGVAENGDSKPDSSKNNPSSRKASSILGKQ 466
Query: 398 E 398
+
Sbjct: 467 Q 467
>At5g10150 unknown protein
Length = 414
Score = 160 bits (406), Expect = 9e-40
Identities = 130/425 (30%), Positives = 211/425 (49%), Gaps = 60/425 (14%)
Query: 1 MSVTSSSRGRPGTRETSPERT-----------KVWTEPKPKTP--RKVSVVYYLSRNGHL 47
M RGR + SPER ++ E K K P R+V VVYYL+RNGHL
Sbjct: 1 MEAVRCRRGRENNK--SPERIIRSLNHHQHDEELEEEVKTKKPIFRRVQVVYYLTRNGHL 58
Query: 48 EHPHFMEV--PLSSPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESD 105
EHPHF+EV P++ P L L+DV+ RL +LRGK M + Y+WS KRSY+NGFVW+DL E+D
Sbjct: 59 EHPHFIEVISPVNQP--LRLRDVMNRLTILRGKCMTSQYAWSCKRSYRNGFVWNDLAEND 116
Query: 106 FIYPTQGHDYILKGSEILE--HETTARPPLE---EESDSPVVITRRRNQSWSSIDLNEYR 160
IYP+ +Y+LKGSEI + E PL +E+ ++ + + ++
Sbjct: 117 VIYPSDCAEYVLKGSEITDKFQEVHVNRPLSGSIQEAPKSRLLRSKLKPQNRTASFDDAE 176
Query: 161 VYKSEPLGE-----NIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETEN 215
+Y E E + + ++ T + R R E E ++K + + E + +
Sbjct: 177 LYVGEEEEEEDGEYELYEEKTSYTSSTTPQSRCSRGVSTETMESTEQKPNLTKTEQDLQV 236
Query: 216 QNQSTELSREEISPPPSDSSPETLGTLMKVDGRLGLRSVPKENPTVESCPSGRMRASSVL 275
++ S++L+R S+P +V R+ ++ VE SGR S+
Sbjct: 237 RSDSSDLTR---------SNPVVKPRRHEVSTRV------EDGDPVEP-GSGR---GSMW 277
Query: 276 LQLLSCGAVSFKEGSGASSG---KDQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEIPDLN 332
LQ++SCG ++ K + + K++ K+ + + + + + + E P
Sbjct: 278 LQMISCGHIATKYYAPSVMNPRQKEENLRKGVLCKNIVKKTVVDDEREMIRFMSENPRFG 337
Query: 333 RVRLEDKEYFSGSLIETKKVE----APAFKRSSSYNADSSSRLQI-----MEHEGDMVRA 383
+ E+KEYFSGS++E+ E P+ +RS+S+N + S +++ + E M +
Sbjct: 338 NPQAEEKEYFSGSIVESVSQERVTAEPSLRRSNSFNEERSKIVEMAKETKKKEERSMAKV 397
Query: 384 KCIPR 388
KCIPR
Sbjct: 398 KCIPR 402
>At1g05577 hypothetical protein
Length = 372
Score = 115 bits (287), Expect = 6e-26
Identities = 82/261 (31%), Positives = 120/261 (45%), Gaps = 44/261 (16%)
Query: 33 RKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSY 92
R+V++VY+LSR+GH++HPH + V S +G++L+DV L RG MP +SWS KR Y
Sbjct: 11 RRVNLVYFLSRSGHVDHPHLLRVHHLSRNGVFLRDVKKWLADARGDAMPDAFSWSCKRRY 70
Query: 93 KNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITRR----RN 148
KNG+VW DL + D I P ++Y+LKGSEIL P E+ +TR
Sbjct: 71 KNGYVWQDLLDDDLITPISDNEYVLKGSEILLSSPKEDYPNVEKK---AWVTRNGGIDAE 127
Query: 149 QSWSSIDLNEYRVYKSEPL--GENIAADASTQTEEKRRRRR--VLREEDEEQEEEEKEKN 204
+ + L ++ K P+ + A ST TEE VL+++D K
Sbjct: 128 EKLQKLKLTSEKIQKESPVFCSQRSTATTSTVTEESTTNEEGFVLKKQD--------PKT 179
Query: 205 DEIENEGETEN-QNQSTELSREEISPPPSDSSPETLGTLMKVDGRLGLRSVPKENPTVES 263
+ +G TEN E R +S S SS +++
Sbjct: 180 VSGQRDGSTENGSGNDVESGRPSVSSTTSSSS------------------------YIKN 215
Query: 264 CPSGRMRASSVLLQLLSCGAV 284
+RAS VL L+ CG +
Sbjct: 216 KSYSSVRASHVLRNLMKCGGL 236
>At1g20290 hypothetical protein
Length = 497
Score = 49.3 bits (116), Expect = 4e-06
Identities = 32/129 (24%), Positives = 54/129 (41%)
Query: 98 WHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLN 157
W + ++SDF T ++ +G+ + E EEE + + +
Sbjct: 337 WVNPSDSDFYKFTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 396
Query: 158 EYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQN 217
E + E E + + EE+ EE+EE+EEEE+E+ +E E E E E +
Sbjct: 397 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 456
Query: 218 QSTELSREE 226
+ E EE
Sbjct: 457 EEEEEEEEE 465
Score = 47.4 bits (111), Expect = 2e-05
Identities = 32/129 (24%), Positives = 51/129 (38%)
Query: 98 WHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLN 157
W L + ++ P+ Y S + E T EEE + + +
Sbjct: 329 WRVLCSTKWVNPSDSDFYKFTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEE 388
Query: 158 EYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQN 217
E + E E + + EE+ EE+EE+EEEE+E+ +E E E E E +
Sbjct: 389 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 448
Query: 218 QSTELSREE 226
+ E EE
Sbjct: 449 EEEEEEEEE 457
Score = 29.3 bits (64), Expect = 4.3
Identities = 21/97 (21%), Positives = 37/97 (37%)
Query: 124 EHETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKR 183
E E EEE + + + E + E E + + EE+
Sbjct: 401 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 460
Query: 184 RRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQST 220
EE+EE+EEEE+E+ + + + + +ST
Sbjct: 461 EEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKKKRRST 497
>At4g08710 putative protein
Length = 715
Score = 45.4 bits (106), Expect = 6e-05
Identities = 29/74 (39%), Positives = 40/74 (53%), Gaps = 8/74 (10%)
Query: 175 ASTQTEE----KRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEI--- 227
A T+TEE ++ V EE EE+E+EE+EK +E E EGE E + + E +EE
Sbjct: 65 APTRTEEPSLTEQDPENVEEEESEEEEKEEEEKEEEEEEEGEEEEEEEEEEEEKEEEENV 124
Query: 228 -SPPPSDSSPETLG 240
SD S +LG
Sbjct: 125 GGEESSDDSTRSLG 138
Score = 40.8 bits (94), Expect = 0.001
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 3/67 (4%)
Query: 165 EPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSR 224
E + E + + + EEK EE+EE+EEEE+EK +E EN G E+ + ST
Sbjct: 80 ENVEEEESEEEEKEEEEKEEEEEEEGEEEEEEEEEEEEKEEE-ENVGGEESSDDSTRSLG 138
Query: 225 EEISPPP 231
+ PPP
Sbjct: 139 KR--PPP 143
Score = 30.0 bits (66), Expect = 2.5
Identities = 18/63 (28%), Positives = 33/63 (51%), Gaps = 5/63 (7%)
Query: 179 TEEKRRRRRVLREEDEEQEEE-EKEKNDEIENEGETENQNQSTEL----SREEISPPPSD 233
+ EK + V++E+D +QEE+ +EK+ + E +G ++ + Q E +E P D
Sbjct: 635 SREKDQEEDVVQEKDGDQEEDVVQEKDGDQEEDGVSKEKEQEKEKDPKEKEKEKDPKEKD 694
Query: 234 SSP 236
P
Sbjct: 695 GGP 697
>At3g02810 putative protein kinase
Length = 558
Score = 44.3 bits (103), Expect = 1e-04
Identities = 32/132 (24%), Positives = 60/132 (45%), Gaps = 10/132 (7%)
Query: 124 EHETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKR 183
EHE P +E + + + SW ID+N+ R+ N D ST +
Sbjct: 434 EHEKDQPPKPIDEKNQAQSLKIKYRYSWEDIDVNDERLSSKSSQKSN---DESTSSRYDS 490
Query: 184 RRRRVLREEDEEQEEEEKEKNDEIEN-EGETENQNQSTELSREEISPPPSDSSPETLGTL 242
R + + ++EE+EEE +EK+ IE+ + + +QS ++ DS + G+L
Sbjct: 491 DRDQDEKGKEEEEEEEAEEKHTHIEHIDSSKTDDDQSVYFDNDD------DSGDDNGGSL 544
Query: 243 MKVDGRLGLRSV 254
++ + + S+
Sbjct: 545 HRIKSDVAIDSI 556
>At5g03710 putative protein
Length = 81
Score = 43.1 bits (100), Expect = 3e-04
Identities = 21/59 (35%), Positives = 31/59 (51%)
Query: 168 GENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
GE + + EE+ EE+EE+EEEE+E+ +E E E E E + + E EE
Sbjct: 5 GEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 63
Score = 42.0 bits (97), Expect = 6e-04
Identities = 19/49 (38%), Positives = 29/49 (58%)
Query: 178 QTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
+ EE+ EE+EE+EEEE+E+ +E E E E E + + + REE
Sbjct: 24 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDREREE 72
Score = 41.6 bits (96), Expect = 8e-04
Identities = 21/60 (35%), Positives = 30/60 (50%)
Query: 178 QTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPE 237
+ EE+ EE+EE+EEEE+E+ +E E E E E + + E EE D E
Sbjct: 12 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDRERE 71
Score = 37.7 bits (86), Expect = 0.012
Identities = 19/68 (27%), Positives = 32/68 (46%)
Query: 151 WSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENE 210
W + E + E E + + EE+ EE+EE+EEEE+E+ +E E E
Sbjct: 4 WGEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 63
Query: 211 GETENQNQ 218
E + + +
Sbjct: 64 EEEDRERE 71
Score = 35.4 bits (80), Expect = 0.060
Identities = 15/42 (35%), Positives = 26/42 (61%)
Query: 178 QTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQS 219
+ EE+ EE+EE+EEEE+E+ +E E E + E + ++
Sbjct: 33 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDREREERA 74
>At5g19310 homeotic gene regulator - like protein
Length = 1041
Score = 40.8 bits (94), Expect = 0.001
Identities = 25/70 (35%), Positives = 38/70 (53%), Gaps = 3/70 (4%)
Query: 174 DASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQN--QSTELSREEISP-P 230
D T+ + VL E DEE+EEEE+E+ +E E E EN+ S + ++++ S P
Sbjct: 962 DTKTRMSNGSKAEAVLSESDEEKEEEEEERKEESGKESEEENEKPLHSWKTNKKKRSRYP 1021
Query: 231 PSDSSPETLG 240
SSP + G
Sbjct: 1022 VMTSSPNSRG 1031
>At1g47300 hypothetical protein
Length = 306
Score = 40.8 bits (94), Expect = 0.001
Identities = 23/70 (32%), Positives = 39/70 (54%), Gaps = 2/70 (2%)
Query: 157 NEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQ 216
+EY Y+S + + AD + + +EE+EE+EEEE+E+ +E E E E E +
Sbjct: 211 HEYDYYRSRVV--YVFADHVEDLDVNDPKLLESKEEEEEEEEEEEEEEEEEEEEEEEEEE 268
Query: 217 NQSTELSREE 226
+S E +E+
Sbjct: 269 EESKEREKEK 278
Score = 35.8 bits (81), Expect = 0.046
Identities = 16/37 (43%), Positives = 25/37 (67%)
Query: 191 EEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEI 227
EE+EE+EEEE+E+ +E E E E E++ + E E +
Sbjct: 247 EEEEEEEEEEEEEEEEEEEEEEEESKEREKEKKIETV 283
Score = 35.8 bits (81), Expect = 0.046
Identities = 15/35 (42%), Positives = 25/35 (70%)
Query: 191 EEDEEQEEEEKEKNDEIENEGETENQNQSTELSRE 225
EE+EE+EEEE+E+ +E E E E E +++ E ++
Sbjct: 245 EEEEEEEEEEEEEEEEEEEEEEEEEESKEREKEKK 279
Score = 32.3 bits (72), Expect = 0.50
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 8/60 (13%)
Query: 154 IDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGET 213
+D+N+ ++ +S+ + + EE+ EE+EE+EEEE+ K E E + ET
Sbjct: 231 LDVNDPKLLESK--------EEEEEEEEEEEEEEEEEEEEEEEEEEEESKEREKEKKIET 282
Score = 30.8 bits (68), Expect = 1.5
Identities = 13/43 (30%), Positives = 26/43 (60%)
Query: 178 QTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQST 220
+++E+ EE+EE+EEEE+E+ +E E++ + + T
Sbjct: 240 ESKEEEEEEEEEEEEEEEEEEEEEEEEEEEESKEREKEKKIET 282
>At5g44800 helicase-like protein
Length = 2228
Score = 40.4 bits (93), Expect = 0.002
Identities = 37/120 (30%), Positives = 50/120 (40%), Gaps = 13/120 (10%)
Query: 117 LKGSEILEHETTARPPLEEES-------DSPVVITRRRNQSWSSIDLNEYRVYKSEPLGE 169
L +E L E PP EE+S ++P + SI L SEP
Sbjct: 2095 LNSTEPLSSEAIIIPPPEEDSVIAAAPSEAPGPSLEGITGTTKSISLESQ---SSEPETI 2151
Query: 170 NIAADASTQTEEKRRRRRVLREEDEEQEEEEKEK--NDEIEN-EGETENQNQSTELSREE 226
N D +T+EK R DE+QEE+E E N + E E E++N N E +E
Sbjct: 2152 NQDGDLDPETDEKVESERTPLHSDEKQEEQESENALNKQCEPIEAESQNTNAEEEAEAQE 2211
>At5g16500 protein kinase-like protein
Length = 636
Score = 40.4 bits (93), Expect = 0.002
Identities = 55/258 (21%), Positives = 103/258 (39%), Gaps = 45/258 (17%)
Query: 129 ARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRV 188
A P+E D + I R+ S S + + + + + +G ++ + ++ EE+ + ++
Sbjct: 362 ATVPMESFRDKSMSIALSRHGSCS---VTPFCISRKD-VGNKSSSSSDSEDEEEEKEQKA 417
Query: 189 LREED------EEQEEEEKEKNDEIENEGETENQNQSTELSR-EEISPPPSDSSPETLGT 241
+EE+ +EQEE + +DE ++ E + + + ++L + E S SDS E
Sbjct: 418 EKEEESTSKKRQEQEETATDSDDESDSNSEKDQEEEQSQLEKARESSSSSSDSGSER--- 474
Query: 242 LMKVDGRLGLRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASSGKDQRFS 301
RS+ + N T +S SS ++ S SS K S
Sbjct: 475 ----------RSIDETNATAQSLKISYSNYSS--------EEEDNEKLSSKSSCKSNEES 516
Query: 302 LVGHYKSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLIETKKVEAPAFKRSS 361
Y S G +H T + I N + +DKE E + E ++
Sbjct: 517 TFSRYDS-----GRDHDDSSRNTSMRI---NSLAHDDKEE-----DEEENHETRSYSDHD 563
Query: 362 SYNADSSSRLQIMEHEGD 379
++S R+ + H+ D
Sbjct: 564 DSPRNTSMRINSLSHDDD 581
>At4g26630 unknown protein
Length = 763
Score = 40.0 bits (92), Expect = 0.002
Identities = 20/44 (45%), Positives = 31/44 (70%), Gaps = 1/44 (2%)
Query: 191 EEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDS 234
EE EE+E++E+EK +E E + E EN+N + S +E +P PS+S
Sbjct: 525 EEKEEEEKQEEEKAEEKEEKKEEENENGIPDKSEDE-APQPSES 567
Score = 35.4 bits (80), Expect = 0.060
Identities = 26/96 (27%), Positives = 40/96 (41%), Gaps = 3/96 (3%)
Query: 169 ENIAADASTQTEEKRRRR--RVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
EN+ EE + + E DE + E+EKE E EN+ E + E +EE
Sbjct: 210 ENVEGKEKEDKEENKTKEVEAAKAEVDESKVEDEKE-GSEDENDNEKVESKDAKEDEKEE 268
Query: 227 ISPPPSDSSPETLGTLMKVDGRLGLRSVPKENPTVE 262
+ D E+ G+ + G V ++N T E
Sbjct: 269 TNDDKEDEKEESKGSKKRGKGTSSGGKVREKNKTEE 304
Score = 32.7 bits (73), Expect = 0.39
Identities = 44/193 (22%), Positives = 76/193 (38%), Gaps = 34/193 (17%)
Query: 110 TQGHDYILKGSEILEH-----ETTARPPLEEESDSPVVITRRRNQSWSSIDL---NEYRV 161
T+ D I K E LE + T + E+ S R+R +S + R
Sbjct: 439 TKKEDIITKLFEFLEKPHVTGDVTGDTTVSEKEKSSKGAKRKRTPKKTSPTAGSSSSKRS 498
Query: 162 YKSEPLGENIA-------ADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIEN----- 209
KS+ E A + ++EE++ ++E+E+ EE+E++K +E EN
Sbjct: 499 AKSQKKSEEATKVVKKSLAHSDDESEEEKEEEE--KQEEEKAEEKEEKKEEENENGIPDK 556
Query: 210 -------EGETENQNQSTELSREEISPPP-----SDSSPETLGTLMKVDGRLGLRSVPKE 257
E+E +++S E S EE + S E+ G + +S P E
Sbjct: 557 SEDEAPQPSESEEKDESEEHSEEETTKKKRGSRLSAGKKESAGRARNKKAVVAAKSSPPE 616
Query: 258 NPTVESCPSGRMR 270
T + + R +
Sbjct: 617 KITQKRSSAKRKK 629
Score = 28.9 bits (63), Expect = 5.6
Identities = 55/253 (21%), Positives = 96/253 (37%), Gaps = 31/253 (12%)
Query: 174 DASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSD 233
D +TE+K + + E Q+ EEK + + E + E N ++ + +E+ +D
Sbjct: 62 DEKAETEDKESEVKKNEDNAETQKMEEKVEVTKDEGQAEATNMDEDADGKKEQ-----TD 116
Query: 234 SSPETLGTLMK--VDGRLG--LRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEG 289
T+MK V+ + + KE + + +A +Q + A K+G
Sbjct: 117 DGVSVEDTVMKENVESKDNNYAKDDEKETKETDITEADHKKAGKEDIQHEADKANGTKDG 176
Query: 290 SGASSGKDQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEIPDLNR--VRLEDKEYFSGSLI 347
+ + + +LV K G ++ VE D N+ +E KE
Sbjct: 177 N--TGDIKEEGTLVDEDK-----------GTDMDEKVENGDENKQVENVEGKEKEDKEEN 223
Query: 348 ETKKVEAPAFKRSSSYNADSSSRLQIMEHEGDMVRAKCIPRKSKTLLTKKEEGTSSSMDS 407
+TK+VEA + S D + E E D + + K +KEE D
Sbjct: 224 KTKEVEAAKAEVDESKVEDEK---EGSEDENDNEKVESKDAKE----DEKEETNDDKEDE 276
Query: 408 VSRSQHGSKRFEG 420
S+ KR +G
Sbjct: 277 KEESKGSKKRGKG 289
Score = 28.9 bits (63), Expect = 5.6
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 5/38 (13%)
Query: 179 TEEKRRRRRVLREE-----DEEQEEEEKEKNDEIENEG 211
T K + +++EE DEE+EEE+KE++ E E G
Sbjct: 716 TPRKSSIKMIIQEELTKLADEEEEEEKKEEDSEKEEAG 753
>At3g14670 hypothetical protein
Length = 232
Score = 40.0 bits (92), Expect = 0.002
Identities = 33/138 (23%), Positives = 56/138 (39%), Gaps = 6/138 (4%)
Query: 126 ETTARPPLEEESDS-----PVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTE 180
+ T PP+ E S PVVI R + + V + E N ++ +
Sbjct: 28 KATTEPPMTTEEPSASKQNPVVIEGRGVEEEQIPTIITTVVEEGEKSDNNEEENSEKDEK 87
Query: 181 EKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPETLG 240
E+ EE EE+E+EE+EK +E G + + S L +E S D +G
Sbjct: 88 EESEEEESEEEEKEEEEKEEEEKEEEGNVAGGGSSDDSSRTLGKESSSDENMDDE-TAVG 146
Query: 241 TLMKVDGRLGLRSVPKEN 258
+ + + + + +EN
Sbjct: 147 KQVDIPAAMKINEMGQEN 164
>At5g57200 putative protein
Length = 591
Score = 39.7 bits (91), Expect = 0.003
Identities = 23/87 (26%), Positives = 39/87 (44%), Gaps = 2/87 (2%)
Query: 178 QTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPE 237
Q+ +++ +E+EEQE+EE++ + E E + EN L EE P + E
Sbjct: 311 QSGSVQKKLEYQEKEEEEQEQEEEQPEEPAEEENQNENTENDQPLIEEEEEEPKEEIEVE 370
Query: 238 TL--GTLMKVDGRLGLRSVPKENPTVE 262
L+ D LGL + + +E
Sbjct: 371 EAKPSPLIDTDDLLGLHEINPKAAEIE 397
>At5g53800 unknown protein
Length = 339
Score = 39.3 bits (90), Expect = 0.004
Identities = 28/108 (25%), Positives = 56/108 (50%), Gaps = 7/108 (6%)
Query: 120 SEILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADAS-TQ 178
SE E + + + +SD +RRR + +SS + SE E+ +D+ ++
Sbjct: 72 SEKEERRRSRKDRGKRKSDRKSSRSRRRRRDYSSSSSD------SESESESEYSDSEESE 125
Query: 179 TEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
+E++RRRR+ R+E EE+E+E K + E + + ++ + +E+
Sbjct: 126 SEDERRRRKRKRKEREEEEKERKRRRREKDKKKRNKSDKDGDKKRKEK 173
>At5g10660 vacuolar calcium binding protein - like
Length = 407
Score = 39.3 bits (90), Expect = 0.004
Identities = 35/129 (27%), Positives = 57/129 (44%), Gaps = 18/129 (13%)
Query: 132 PLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLRE 191
P EE+ D + ++S + + E GE I D ST+ E+ + + E
Sbjct: 225 PKEEDKDQ----FAQPDESGEEKETSPVAASTEEQKGELIDEDKSTEQIEEPKEPENIEE 280
Query: 192 EDEEQEEEEKEKNDEIENE---------GETENQNQSTELSRE--EISPPPSDSS---PE 237
+ E+EEE K+K+D+ EN E N +S E +E E+ +SS E
Sbjct: 281 NNSEEEEEVKKKSDDEENSETVATTTDMNEAVNVEESKEEEKEEAEVKEEEGESSAAKEE 340
Query: 238 TLGTLMKVD 246
T T+ +V+
Sbjct: 341 TTETMAQVE 349
Score = 32.3 bits (72), Expect = 0.50
Identities = 31/117 (26%), Positives = 50/117 (42%), Gaps = 16/117 (13%)
Query: 134 EEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLG-ENIAADASTQTEEKRRR------- 185
EE+ SPV + Q ID ++ EP ENI + S + EE +++
Sbjct: 241 EEKETSPVAASTEE-QKGELIDEDKSTEQIEEPKEPENIEENNSEEEEEVKKKSDDEENS 299
Query: 186 RRVLREED-------EEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSS 235
V D EE +EEEKE+ + E EGE+ + T + ++ P + +
Sbjct: 300 ETVATTTDMNEAVNVEESKEEEKEEAEVKEEEGESSAAKEETTETMAQVEELPEEGT 356
>At3g27720 hypothetical protein
Length = 493
Score = 39.3 bits (90), Expect = 0.004
Identities = 41/168 (24%), Positives = 73/168 (43%), Gaps = 10/168 (5%)
Query: 103 ESDFIYPTQ--GHDYILKGSEILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYR 160
E D YP++ HD + +E E+ + E S ++ NQ+ + + ++
Sbjct: 11 EEDNCYPSEFDDHDQMCSNAE----ESDLQHSREPTSQVMEFLSVTENQARTLLIQYQWN 66
Query: 161 VYKSEPLGENIAADAS-TQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQS 219
V K + + D ++ EE+ + E E++EEEE +++E E GE E +
Sbjct: 67 VDKLFSVYTDQGKDVLFSRAEEEEGEDKEEEEGGEDEEEEEGGEDEEAEEGGEDEEAEEG 126
Query: 220 TELSREEISPPPSDSSPETLGT-LMKVDGRLGLRSVPKENPTVESCPS 266
E EE D + + T L + R + S ++N V+ CPS
Sbjct: 127 VEDEEEE--EDEKDDEVQLVSTELAEKFDRFLIESYVEDNNMVKWCPS 172
>At1g66660 hypothetical protein
Length = 412
Score = 39.3 bits (90), Expect = 0.004
Identities = 30/88 (34%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 191 EEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPETLGTLMKVDGRLG 250
EEDEE+ EEE+E+ +E E+E E E +N +T +E S P S P L + +D
Sbjct: 118 EEDEEEFEEEEEELEEEEDEEEEEEENVTT----DEQSGSPKSSQPVKLQSSDVLD--CP 171
Query: 251 LRSVPKENPTVESCPSGRMRASSVLLQL 278
P + P + C +G + SS +L
Sbjct: 172 TCCEPLKRP-IYQCSNGHLACSSCCQKL 198
Score = 30.8 bits (68), Expect = 1.5
Identities = 15/63 (23%), Positives = 29/63 (45%)
Query: 174 DASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSD 233
D + +E+ E+EE EEEE+E+N + + + +Q +L ++ P+
Sbjct: 114 DGEFEEDEEEFEEEEEELEEEEDEEEEEEENVTTDEQSGSPKSSQPVKLQSSDVLDCPTC 173
Query: 234 SSP 236
P
Sbjct: 174 CEP 176
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.128 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,460,025
Number of Sequences: 26719
Number of extensions: 514903
Number of successful extensions: 8597
Number of sequences better than 10.0: 493
Number of HSP's better than 10.0 without gapping: 293
Number of HSP's successfully gapped in prelim test: 216
Number of HSP's that attempted gapping in prelim test: 4343
Number of HSP's gapped (non-prelim): 1969
length of query: 422
length of database: 11,318,596
effective HSP length: 102
effective length of query: 320
effective length of database: 8,593,258
effective search space: 2749842560
effective search space used: 2749842560
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC139841.8


