

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.2 - phase: 0 /pseudo
(285 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
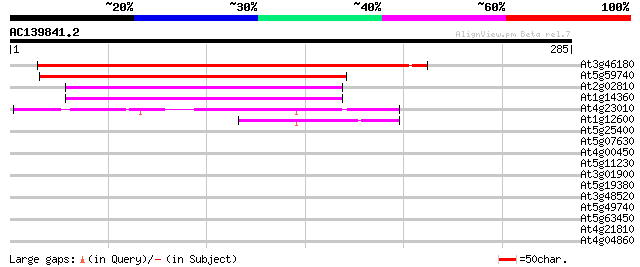
Score E
Sequences producing significant alignments: (bits) Value
At3g46180 unknown protein 305 1e-83
At5g59740 protein serine /threonine kinase - like protein 256 1e-68
At2g02810 unknown protein 77 1e-14
At1g14360 putative UDP-galactose transporter MSS4 75 4e-14
At4g23010 unknown protein 49 3e-06
At1g12600 hypothetical protein 42 4e-04
At5g25400 putative protein 33 0.23
At5g07630 putative protein 32 0.30
At4g00450 hypothetical protein 32 0.51
At5g11230 putative protein 30 1.1
At3g01900 putative cytochrome P450 30 1.9
At5g19380 unknown protein 29 3.3
At3g48520 cytochrome P450-like protein 29 3.3
At5g49740 FRO1-like protein; NADPH oxidase-like 28 4.3
At5g63450 cytochrome P450-like protein 28 5.6
At4g21810 unknown protein 28 5.6
At4g04860 unknown protein 27 9.6
>At3g46180 unknown protein
Length = 347
Score = 305 bits (782), Expect = 1e-83
Identities = 146/198 (73%), Positives = 172/198 (86%), Gaps = 1/198 (0%)
Query: 15 SLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRI 74
S++++ + KLWK FA+SGIMLTLV YG+LQEKIMRVPYG+ K+YFK+SLFLVFCNR+
Sbjct: 6 SVNEAKEKKKKLWKAVFAISGIMLTLVIYGLLQEKIMRVPYGLKKEYFKHSLFLVFCNRL 65
Query: 75 TTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKM 134
TTSAVSAAAL+A K L PVAP+YKYCL+SV+NILTTTCQYEALKYVSFPVQTLAKCAKM
Sbjct: 66 TTSAVSAAALLASKKVLDPVAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKM 125
Query: 135 IPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMI 194
IPVM+W LIMQKKY+G DYL+AF VTLGCS+FIL+PAG DISPY +GRENT+WG+ LM+
Sbjct: 126 IPVMVWGTLIMQKKYRGFDYLVAFLVTLGCSVFILFPAGDDISPYNKGRENTVWGVSLMV 185
Query: 195 GYLGYHYKTQSLFAFRLF 212
GYLG+ T S F +LF
Sbjct: 186 GYLGFDGFT-STFQDKLF 202
>At5g59740 protein serine /threonine kinase - like protein
Length = 295
Score = 256 bits (653), Expect = 1e-68
Identities = 124/156 (79%), Positives = 138/156 (87%)
Query: 16 LDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRIT 75
L + V NKLWK FAVSGIM TLV YGVLQEKIMRVPYGVNK++FK+SLFLVFCNR+T
Sbjct: 6 LVNGGVKENKLWKGVFAVSGIMSTLVIYGVLQEKIMRVPYGVNKEFFKHSLFLVFCNRLT 65
Query: 76 TSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMI 135
TSAVSA AL+A K L PVAP+YKYCL+SV+NILTTTCQYEALKYVSFPVQTLAKCAKMI
Sbjct: 66 TSAVSAGALLASKKVLDPVAPVYKYCLISVTNILTTTCQYEALKYVSFPVQTLAKCAKMI 125
Query: 136 PVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYP 171
PVM+W LIMQKKY+G DYL+AF VTLGCS+FIL+P
Sbjct: 126 PVMVWGTLIMQKKYKGFDYLVAFLVTLGCSVFILFP 161
>At2g02810 unknown protein
Length = 332
Score = 76.6 bits (187), Expect = 1e-14
Identities = 46/141 (32%), Positives = 69/141 (48%)
Query: 29 VAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRITTSAVSAAALVARD 88
+A +SGI + GVLQE + +G ++ F++ FL + S +
Sbjct: 14 LALCISGIWSAYIYQGVLQETLSTKRFGPDEKRFEHLAFLNLAQSVVCLIWSYIMIKLWS 73
Query: 89 KALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKK 148
A AP + Y ++N + EALKY+S+P Q LAK +KMIPVM+ L+ +
Sbjct: 74 NAGNGGAPWWTYWSAGITNTIGPAMGIEALKYISYPAQVLAKSSKMIPVMLMGTLVYGIR 133
Query: 149 YQGTDYLLAFSVTLGCSIFIL 169
Y +Y+ F V G SIF L
Sbjct: 134 YTFPEYMCTFLVAGGVSIFAL 154
>At1g14360 putative UDP-galactose transporter MSS4
Length = 331
Score = 75.1 bits (183), Expect = 4e-14
Identities = 44/141 (31%), Positives = 69/141 (48%)
Query: 29 VAFAVSGIMLTLVTYGVLQEKIMRVPYGVNKDYFKYSLFLVFCNRITTSAVSAAALVARD 88
++F V+GI + G+LQE + +G + F++ FL + S +
Sbjct: 14 LSFCVAGIWAAYIYQGILQETLSTKKFGEDGKRFEHLAFLNLAQNVICLVWSYIMIKLWS 73
Query: 89 KALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKK 148
AP + Y ++N + EALKY+S+P Q LAK +KMIPVM+ +L+ +
Sbjct: 74 NGGSGGAPWWTYWSAGITNTIGPAMGIEALKYISYPAQVLAKSSKMIPVMLMGSLVYGIR 133
Query: 149 YQGTDYLLAFSVTLGCSIFIL 169
Y +YL F V G S+F L
Sbjct: 134 YTLPEYLCTFLVAGGVSMFAL 154
>At4g23010 unknown protein
Length = 345
Score = 48.9 bits (115), Expect = 3e-06
Identities = 55/205 (26%), Positives = 87/205 (41%), Gaps = 30/205 (14%)
Query: 3 MAETTTSSSSSSSLDDSNVTRNKLWKVAFAVSGIMLTLVTYGVLQEKIM-RVPYGVNKDY 61
M E T S SL D + L SG + GV +E + R+ + Y
Sbjct: 1 MKEEQTRSLFGISLSDKPTWQQFL----ICTSGFFFGYLVNGVCEEYVYNRLQFSFGW-Y 55
Query: 62 FKYS-----LFLVFCNRITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYE 116
F + LFL++ TT + V P+ Y +S + +
Sbjct: 56 FTFIQGFVYLFLIYLQGFTTKHI--------------VNPMRTYVKLSAVLMGSHGLTKG 101
Query: 117 ALKYVSFPVQTLAKCAKMIPVMIWSALI--MQKKYQGTDYLLAFSVTLGCSIFILYPAGT 174
+L Y+++P Q + K K++PVMI A I +++KY +Y+ AF + LG +F L A
Sbjct: 102 SLAYLNYPAQIMFKSTKVLPVMIMGAFIPGLRRKYPVHEYISAFLLVLGLILFTL--ADA 159
Query: 175 DISP-YGRGRENTIWGILLMIGYLG 198
+SP + I G L+M +LG
Sbjct: 160 QMSPNFSMIGIMMITGALIMDAFLG 184
>At1g12600 hypothetical protein
Length = 155
Score = 42.0 bits (97), Expect = 4e-04
Identities = 26/84 (30%), Positives = 46/84 (53%), Gaps = 3/84 (3%)
Query: 117 ALKYVSFPVQTLAKCAKMIPVMIWSALI--MQKKYQGTDYLLAFSVTLGCSIFILYPAGT 174
+L Y+++P Q + K K++PVM+ A I +++KY +Y+ A + +G +F L A T
Sbjct: 33 SLAYLNYPAQIMFKSTKVLPVMVMGAFIPGLRRKYPVHEYISAMLLVIGLILFTLADAHT 92
Query: 175 DISPYGRGRENTIWGILLMIGYLG 198
+ + I G L+M +LG
Sbjct: 93 SPN-FSIIGVMMISGALIMDAFLG 115
>At5g25400 putative protein
Length = 349
Score = 32.7 bits (73), Expect = 0.23
Identities = 40/198 (20%), Positives = 83/198 (41%), Gaps = 32/198 (16%)
Query: 36 IMLTLVTYG--VLQEKIMRVPYGVNKDYFKYSLFLVFCNRITTSAVSAAALV-----ARD 88
+ T++ Y +L +K+ P+ ++ S FC+ + + V +RD
Sbjct: 28 LSFTVIVYNKYILDKKMYDWPFPISLTMIHMS----FCSTLAFLLIKVFKFVEPVSMSRD 83
Query: 89 KALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKK 148
L V PI L S+S L+ + Y+ V + ++PV ++S ++ KK
Sbjct: 84 TYLRSVVPIG--ALYSLSLWLSNSA------YIYLSVSFIQMLKALMPVAVYSIGVLFKK 135
Query: 149 --YQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKTQSL 206
++ + S++ G +I + YG R + +WG++L +G + + +
Sbjct: 136 EGFKSETMMNMLSISFGVAI----------AAYGEARFD-VWGVILQLGAVAFEATRLVM 184
Query: 207 FAFRLFSLAYFVSFLSSL 224
L S ++ ++SL
Sbjct: 185 IQILLTSKGITLNPITSL 202
>At5g07630 putative protein
Length = 401
Score = 32.3 bits (72), Expect = 0.30
Identities = 32/123 (26%), Positives = 50/123 (40%), Gaps = 26/123 (21%)
Query: 108 ILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIF 167
IL+ T L+ + V TLA+C V ++S L+MQ + V G S+F
Sbjct: 75 ILSQTLLKLQLRLIVETVATLARC-----VTLYSLLVMQTNMEKVIIFALSQVAYGGSLF 129
Query: 168 ILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKTQSLFAFRLFSLAYFVSFLSSLLES 227
I Y W L+ G Y++ +LF FR + F + LS + +
Sbjct: 130 IGY-----------------WAYFLICGV----YRSSNLFPFRPGNFMDFDNDLSKMCKL 168
Query: 228 HSF 230
+F
Sbjct: 169 FTF 171
>At4g00450 hypothetical protein
Length = 2124
Score = 31.6 bits (70), Expect = 0.51
Identities = 32/140 (22%), Positives = 67/140 (47%), Gaps = 10/140 (7%)
Query: 5 ETTTSSSSSSSLDDSNVTR----NKLWKVAFAVSGIMLTLVTYGVLQ-EKIMRVPYGVNK 59
E+ S++++S++D SN T ++ K+ AVS +++ +T+GV+ E+I+ + K
Sbjct: 1299 ESYDSNANNSTIDMSNGTGKMALSRATKITAAVSALVIGSITHGVITLERIVGLLR--LK 1356
Query: 60 DYFKYSLFLVFCNRITTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALK 119
DY F+ F R +S+ +A + K P+ + + V N T +
Sbjct: 1357 DYLD---FVQFVRRTKSSSNGSARSMGASKVESPIEVYVHWFRLLVGNCKTVSEGLVLEL 1413
Query: 120 YVSFPVQTLAKCAKMIPVMI 139
V +++ +M+P+ +
Sbjct: 1414 VGESSVVAISRMQRMLPLKL 1433
>At5g11230 putative protein
Length = 351
Score = 30.4 bits (67), Expect = 1.1
Identities = 33/186 (17%), Positives = 75/186 (39%), Gaps = 24/186 (12%)
Query: 20 NVTRNKLWKVAFAVSGIML--TLVTYG--VLQEKIMRVPYGVNKDYFKYSLFLVFCNRIT 75
+V +N + ++ I L T++ Y +L +K+ P+ ++ S FC+ +
Sbjct: 10 SVIKNIVLSYSYVAIWIFLSFTVIVYNKYILDKKMYNWPFPISLTMIHMS----FCSTLA 65
Query: 76 TSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMI 135
+ V K Y +V + + + Y+ V + ++
Sbjct: 66 FLIIKVFKFVEPVKM---TRETYLRSVVPIGALYALSLWLSNSAYIYLSVSFIQMLKALM 122
Query: 136 PVMIWSALIMQKK--YQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLM 193
PV ++S ++ KK ++ + S++ G +I + YG R + +WG++L
Sbjct: 123 PVAVYSIGVLFKKEGFKSDTMMNMLSISFGVAI----------AAYGEARFD-VWGVILQ 171
Query: 194 IGYLGY 199
+G + +
Sbjct: 172 LGAVAF 177
>At3g01900 putative cytochrome P450
Length = 496
Score = 29.6 bits (65), Expect = 1.9
Identities = 23/86 (26%), Positives = 35/86 (39%), Gaps = 11/86 (12%)
Query: 104 SVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLG 163
SV +T YE+LK +S L + ++ P + W + K+ TD L +
Sbjct: 323 SVKEEITGGFDYESLKKLSLLKACLCEVMRLYPPVPWDS-----KHALTDDRLPDGTLVR 377
Query: 164 CSIFILYPAGTDISPYGRGRENTIWG 189
+ Y PYG GR +WG
Sbjct: 378 AGDRVTY------FPYGMGRMEELWG 397
>At5g19380 unknown protein
Length = 447
Score = 28.9 bits (63), Expect = 3.3
Identities = 37/156 (23%), Positives = 63/156 (39%), Gaps = 14/156 (8%)
Query: 75 TTSAVSAAALVARDKALLPVAPIYKYCLVSVSNILTTTCQYEALKYVSFPVQTLAKCAKM 134
TT+A+ V AL+P+ Y + L +S Y ++ Y + + K +M
Sbjct: 111 TTAALGVGNRVLYKLALIPLKQ-YPFFLAQLSTF-GYVAVYFSILYFRYRAGIVTK--EM 166
Query: 135 IPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMI 194
+ V LI+ G LA + + + + P+ T +S + +W IL I
Sbjct: 167 LSVPKLPFLIV-----GVLESLALAAGMAAASNLSGPSTTVLS-----QTFLVWQILFSI 216
Query: 195 GYLGYHYKTQSLFAFRLFSLAYFVSFLSSLLESHSF 230
+LG Y+ + L + VS S +HSF
Sbjct: 217 IFLGRRYRINQILGCTLVAFGVIVSVASGSGAAHSF 252
>At3g48520 cytochrome P450-like protein
Length = 506
Score = 28.9 bits (63), Expect = 3.3
Identities = 20/88 (22%), Positives = 37/88 (41%), Gaps = 11/88 (12%)
Query: 102 LVSVSNILTTTCQYEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVT 161
L V +++ +E LK +++ L + ++ P + W + K+ D +L
Sbjct: 330 LEEVDPLVSLGLGFEDLKEMAYTKACLCEAMRLYPPVSWDS-----KHAANDDVLPDGTR 384
Query: 162 LGCSIFILYPAGTDISPYGRGRENTIWG 189
+ + Y PYG GR T+WG
Sbjct: 385 VKRGDKVTY------FPYGMGRMETLWG 406
>At5g49740 FRO1-like protein; NADPH oxidase-like
Length = 739
Score = 28.5 bits (62), Expect = 4.3
Identities = 24/89 (26%), Positives = 44/89 (48%), Gaps = 8/89 (8%)
Query: 2 VMAETTTSSSSSSSLDDSNVTRNKLWKVAFAVSGIMLTLVTY-----GVLQEKIMRVPYG 56
++++ +SSSSSSS S V + W + +S I +T V + G L ++I+
Sbjct: 8 LLSKDLSSSSSSSSSSSSVVVSSLKWILKVVMSVIFVTWVVFLMMYPGSLGDQILTNWRA 67
Query: 57 VNKDY---FKYSLFLVFCNRITTSAVSAA 82
++ + S+FL+F I A+ A+
Sbjct: 68 ISSNTLFGLTGSMFLIFSGPILVIAILAS 96
>At5g63450 cytochrome P450-like protein
Length = 510
Score = 28.1 bits (61), Expect = 5.6
Identities = 18/75 (24%), Positives = 31/75 (41%), Gaps = 11/75 (14%)
Query: 115 YEALKYVSFPVQTLAKCAKMIPVMIWSALIMQKKYQGTDYLLAFSVTLGCSIFILYPAGT 174
+E L+ +S+ L + ++ P + W + K+ D +L L + Y
Sbjct: 345 FEDLREMSYTKACLCEAMRLYPPVAWDS-----KHAANDDILPDGTPLKKGDKVTY---- 395
Query: 175 DISPYGRGRENTIWG 189
PYG GR +WG
Sbjct: 396 --FPYGMGRMEKVWG 408
>At4g21810 unknown protein
Length = 244
Score = 28.1 bits (61), Expect = 5.6
Identities = 26/78 (33%), Positives = 38/78 (48%), Gaps = 8/78 (10%)
Query: 154 YLLAFSVT-LGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKTQSLFAFRLF 212
YL A VT +GCS+ I+ P ++P ++ W L+ +L Y K F F +F
Sbjct: 19 YLTAAVVTTVGCSLEIISPYNLYLNPTLVVKQYQFW--RLVTNFL-YFRKMDLDFLFHMF 75
Query: 213 SLAYFVSFLSSLLESHSF 230
LA + LLE +SF
Sbjct: 76 FLARY----CKLLEENSF 89
>At4g04860 unknown protein
Length = 244
Score = 27.3 bits (59), Expect = 9.6
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 7/74 (9%)
Query: 157 AFSVTLGCSIFILYPAGTDISPYGRGRENTIWGILLMIGYLGYHYKTQSLFAFRLFSLAY 216
A T+GCS+ I+ P ++P ++ W L+ +L Y K F F +F LA
Sbjct: 23 AVITTVGCSLDIISPYNLYLNPTLVVKQYQYW--RLVTNFL-YFRKMDLDFMFHMFFLAR 79
Query: 217 FVSFLSSLLESHSF 230
+ LLE +SF
Sbjct: 80 Y----CKLLEENSF 89
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.334 0.143 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,409,965
Number of Sequences: 26719
Number of extensions: 199213
Number of successful extensions: 898
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 886
Number of HSP's gapped (non-prelim): 17
length of query: 285
length of database: 11,318,596
effective HSP length: 98
effective length of query: 187
effective length of database: 8,700,134
effective search space: 1626925058
effective search space used: 1626925058
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC139841.2


