

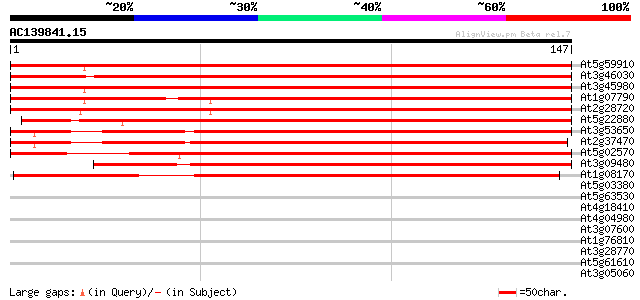
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.15 - phase: 0
(147 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59910 histone H2B - like protein 250 2e-67
At3g46030 histone H2B -like protein 250 2e-67
At3g45980 histone H2B 248 1e-66
At1g07790 histone H2B 241 8e-65
At2g28720 putative histone H2B 240 2e-64
At5g22880 histone H2B like protein (emb|CAA69025.1) 235 6e-63
At3g53650 histone H2B - like protein 228 9e-61
At2g37470 putative histone H2B 220 2e-58
At5g02570 putative protein 213 3e-56
At3g09480 putative histone H2B 204 1e-53
At1g08170 hypothetical protein 130 2e-31
At5g03380 farnesylated protein - like 38 0.002
At5g63530 putative metal-binding protein 36 0.006
At4g18410 MuDR transposable element - like protein 35 0.019
At4g04980 34 0.032
At3g07600 unknown protein 34 0.032
At1g76810 translation initiation factor IF-2 like protein 33 0.042
At3g28770 hypothetical protein 33 0.055
At5g61610 putative protein 33 0.071
At3g05060 putative SAR DNA-binding protein-1 33 0.071
>At5g59910 histone H2B - like protein
Length = 150
Score = 250 bits (638), Expect = 2e-67
Identities = 130/150 (86%), Positives = 136/150 (90%), Gaps = 3/150 (2%)
Query: 1 MAPKGEKKPAEKKPAEEK---KSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNV 57
MAPK EKKPAEKKPA EK + + AEKAPAEKKPKAGKKLPKE G+ KKK KK+V
Sbjct: 1 MAPKAEKKPAEKKPASEKPVEEKSKAEKAPAEKKPKAGKKLPKEAGAGGDKKKKMKKKSV 60
Query: 58 ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREI 117
ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQE+S+LARYNKKPTITSREI
Sbjct: 61 ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQEASKLARYNKKPTITSREI 120
Query: 118 QTAVRLVLPGELAKHAVSEGTKAVTKFTSS 147
QTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 121 QTAVRLVLPGELAKHAVSEGTKAVTKFTSS 150
>At3g46030 histone H2B -like protein
Length = 145
Score = 250 bits (638), Expect = 2e-67
Identities = 130/147 (88%), Positives = 133/147 (90%), Gaps = 2/147 (1%)
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVETY 60
MAPK EKKPAEKKP EEK AEKAPAEKKPKAGKKLPKE G+ KKK KK+VETY
Sbjct: 1 MAPKAEKKPAEKKPVEEKSK--AEKAPAEKKPKAGKKLPKEAGAGGDKKKKMKKKSVETY 58
Query: 61 KIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTA 120
KIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLA ESS+LARYNKKPTITSREIQTA
Sbjct: 59 KIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLASESSKLARYNKKPTITSREIQTA 118
Query: 121 VRLVLPGELAKHAVSEGTKAVTKFTSS 147
VRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 119 VRLVLPGELAKHAVSEGTKAVTKFTSS 145
>At3g45980 histone H2B
Length = 150
Score = 248 bits (632), Expect = 1e-66
Identities = 129/150 (86%), Positives = 135/150 (90%), Gaps = 3/150 (2%)
Query: 1 MAPKGEKKPAEKKPAEEK---KSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNV 57
MAP+ EKKPAEKKPA EK + + AEKAPAEKKPKAGKKLPKE G+ KKK KK+V
Sbjct: 1 MAPRAEKKPAEKKPAAEKPVEEKSKAEKAPAEKKPKAGKKLPKEAGAGGDKKKKMKKKSV 60
Query: 58 ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREI 117
ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLA ESS+LARYNKKPTITSREI
Sbjct: 61 ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLASESSKLARYNKKPTITSREI 120
Query: 118 QTAVRLVLPGELAKHAVSEGTKAVTKFTSS 147
QTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 121 QTAVRLVLPGELAKHAVSEGTKAVTKFTSS 150
>At1g07790 histone H2B
Length = 148
Score = 241 bits (616), Expect = 8e-65
Identities = 131/151 (86%), Positives = 137/151 (89%), Gaps = 7/151 (4%)
Query: 1 MAPKGEKKPAEKKPAEEK---KSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKK-RSKKN 56
MAP+ EKKPAEKK A E+ ++ AEKAPAEKKPKAGKKLP + AGDKKK RSKKN
Sbjct: 1 MAPRAEKKPAEKKTAAERPVEENKAAEKAPAEKKPKAGKKLPPK---EAGDKKKKRSKKN 57
Query: 57 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 116
VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESS+LARYNKKPTITSRE
Sbjct: 58 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSKLARYNKKPTITSRE 117
Query: 117 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 147
IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 118 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
>At2g28720 putative histone H2B
Length = 151
Score = 240 bits (612), Expect = 2e-64
Identities = 128/151 (84%), Positives = 136/151 (89%), Gaps = 4/151 (2%)
Query: 1 MAPKGEKKPAEKKPAEE---KKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKK-RSKKN 56
MAPK KKPAEKKPAE+ ++ VAEKAPAEKKPKAGKKLPKE + +KKK R KK+
Sbjct: 1 MAPKAGKKPAEKKPAEKAPAEEEKVAEKAPAEKKPKAGKKLPKEAVTGGVEKKKKRVKKS 60
Query: 57 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 116
ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQE+S+LARYNKKPTITSRE
Sbjct: 61 TETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQEASKLARYNKKPTITSRE 120
Query: 117 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 147
IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 121 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 151
>At5g22880 histone H2B like protein (emb|CAA69025.1)
Length = 145
Score = 235 bits (600), Expect = 6e-63
Identities = 127/145 (87%), Positives = 130/145 (89%), Gaps = 3/145 (2%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPA-EKKPKAGKKLPKEGGSAAGDKKKRSKKNVETYKI 62
K +KKPAEKKPAE K+ AE A A EKKPKAGKKLPKE A KKKRSKKNVETYKI
Sbjct: 3 KADKKPAEKKPAE--KTPAAEPAAAAEKKPKAGKKLPKEPAGAGDKKKKRSKKNVETYKI 60
Query: 63 YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAVR 122
YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLA ESS+LARYNKKPTITSREIQTAVR
Sbjct: 61 YIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAGESSKLARYNKKPTITSREIQTAVR 120
Query: 123 LVLPGELAKHAVSEGTKAVTKFTSS 147
LVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 121 LVLPGELAKHAVSEGTKAVTKFTSS 145
>At3g53650 histone H2B - like protein
Length = 138
Score = 228 bits (581), Expect = 9e-61
Identities = 125/148 (84%), Positives = 129/148 (86%), Gaps = 11/148 (7%)
Query: 1 MAPKG-EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVET 59
MAPK EKKPA KKPAE KAPAEK PKA KK+ KEGGS KKK+SKKN+ET
Sbjct: 1 MAPKAAEKKPAGKKPAE--------KAPAEKLPKAEKKITKEGGSEK--KKKKSKKNIET 50
Query: 60 YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 119
YKIYIFKVLKQVHPDIGIS KAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT
Sbjct: 51 YKIYIFKVLKQVHPDIGISGKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 110
Query: 120 AVRLVLPGELAKHAVSEGTKAVTKFTSS 147
AVRLVLPGEL+KHAVSEGTKAVTKFTSS
Sbjct: 111 AVRLVLPGELSKHAVSEGTKAVTKFTSS 138
>At2g37470 putative histone H2B
Length = 138
Score = 220 bits (561), Expect = 2e-58
Identities = 120/147 (81%), Positives = 127/147 (85%), Gaps = 10/147 (6%)
Query: 1 MAPKG-EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVET 59
MAPK EKKPAEKKPA KAPAEK PKA KK+ K+ G + KKK+SKK+VET
Sbjct: 1 MAPKAAEKKPAEKKPAG--------KAPAEKLPKAEKKISKDAGGSE-KKKKKSKKSVET 51
Query: 60 YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 119
YKIYIFKVLKQVHPD+GIS KAMGIMNSFINDIFEKLAQESS+LARYNKKPTITSREIQT
Sbjct: 52 YKIYIFKVLKQVHPDVGISGKAMGIMNSFINDIFEKLAQESSKLARYNKKPTITSREIQT 111
Query: 120 AVRLVLPGELAKHAVSEGTKAVTKFTS 146
AVRLVLPGELAKHAVSEGTKAVTKFTS
Sbjct: 112 AVRLVLPGELAKHAVSEGTKAVTKFTS 138
>At5g02570 putative protein
Length = 132
Score = 213 bits (542), Expect = 3e-56
Identities = 117/148 (79%), Positives = 121/148 (81%), Gaps = 17/148 (11%)
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGG-SAAGDKKKRSKKNVET 59
MAPK EKKPAEK PA PKA KK+ KEGG S KKK++KK+ ET
Sbjct: 1 MAPKAEKKPAEKAPA----------------PKAEKKIAKEGGTSEIVKKKKKTKKSTET 44
Query: 60 YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 119
YKIYIFKVLKQVHPDIGIS KAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT
Sbjct: 45 YKIYIFKVLKQVHPDIGISGKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 104
Query: 120 AVRLVLPGELAKHAVSEGTKAVTKFTSS 147
AVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 105 AVRLVLPGELAKHAVSEGTKAVTKFTSS 132
>At3g09480 putative histone H2B
Length = 126
Score = 204 bits (520), Expect = 1e-53
Identities = 105/125 (84%), Positives = 114/125 (91%), Gaps = 3/125 (2%)
Query: 23 AEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVETYKIYIFKVLKQVHPDIGISSKAM 82
AEK P+EK PKA KK+ KEGGS ++KK++KK+ ETYKIY+FKVLKQVHPDIGIS KAM
Sbjct: 5 AEKKPSEKAPKADKKITKEGGS---ERKKKTKKSTETYKIYLFKVLKQVHPDIGISGKAM 61
Query: 83 GIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAVRLVLPGELAKHAVSEGTKAVT 142
GIMNSFIND FEK+A ESSRLARYNKKPTITSREIQTAVRLVLPGELAKHAVSEGTKAVT
Sbjct: 62 GIMNSFINDTFEKIALESSRLARYNKKPTITSREIQTAVRLVLPGELAKHAVSEGTKAVT 121
Query: 143 KFTSS 147
KFTSS
Sbjct: 122 KFTSS 126
>At1g08170 hypothetical protein
Length = 243
Score = 130 bits (328), Expect = 2e-31
Identities = 67/143 (46%), Positives = 96/143 (66%), Gaps = 14/143 (9%)
Query: 2 APKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVETYK 61
+P+ + PA K KK+ EK KK K KKKR + Y+
Sbjct: 107 SPQPPETPASKSEGTLKKTDKVEKKQENKKKKK--------------KKKRDDLAGDEYR 152
Query: 62 IYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAV 121
Y++KV+KQVHPD+GI+SKAM ++N F+ D+FE++AQE++RL+ Y K+ T++SREI+ AV
Sbjct: 153 RYVYKVMKQVHPDLGITSKAMTVVNMFMGDMFERIAQEAARLSDYTKRRTLSSREIEAAV 212
Query: 122 RLVLPGELAKHAVSEGTKAVTKF 144
RLVLPGEL++HAV+EG+KAV+ F
Sbjct: 213 RLVLPGELSRHAVAEGSKAVSNF 235
>At5g03380 farnesylated protein - like
Length = 392
Score = 37.7 bits (86), Expect = 0.002
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKK 55
+AP ++ P AE+K S AE+ PAEKKP A +K G+KK+ KK
Sbjct: 92 VAPPKKETPPSSGGAEKKPSPAAEEKPAEKKPAAVEK--------PGEKKEEKKK 138
Score = 26.6 bits (57), Expect = 5.1
Identities = 14/37 (37%), Positives = 20/37 (53%), Gaps = 6/37 (16%)
Query: 2 APKGEKKPAEKKPA------EEKKSTVAEKAPAEKKP 32
+P E+KPAEKKPA E+K+ E+ + P
Sbjct: 111 SPAAEEKPAEKKPAAVEKPGEKKEEKKKEEGEKKASP 147
>At5g63530 putative metal-binding protein
Length = 355
Score = 36.2 bits (82), Expect = 0.006
Identities = 22/45 (48%), Positives = 26/45 (56%), Gaps = 1/45 (2%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKK 50
EKK EKKP EEKK +K AEKK + K P+EG S K+
Sbjct: 13 EKKMEEKKP-EEKKEGEDKKVDAEKKGEDSDKKPQEGESNKDSKE 56
Score = 30.8 bits (68), Expect = 0.27
Identities = 16/40 (40%), Positives = 20/40 (50%)
Query: 11 EKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKK 50
EKKP ++ + EK P EKK KK+ E DKK
Sbjct: 5 EKKPEAAEEKKMEEKKPEEKKEGEDKKVDAEKKGEDSDKK 44
>At4g18410 MuDR transposable element - like protein
Length = 633
Score = 34.7 bits (78), Expect = 0.019
Identities = 24/79 (30%), Positives = 38/79 (47%), Gaps = 5/79 (6%)
Query: 3 PKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVETYKI 62
P+GE+ K AE+K+ ++P +K+PKA K++ G A D R KN +
Sbjct: 560 PEGEEDKKMTK-AEKKRKKGVNESPTKKQPKAKKRIMHCGVCGAADHNSRHHKNDKVSS- 617
Query: 63 YIFKVLKQVHPDIGISSKA 81
L Q+ P G ++A
Sbjct: 618 ---SQLSQLEPSQGSLTQA 633
>At4g04980
Length = 681
Score = 33.9 bits (76), Expect = 0.032
Identities = 32/124 (25%), Positives = 55/124 (43%), Gaps = 15/124 (12%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEG-GSAAGDKKKRS------KKN 56
+G + K A + +++VAEK+P K+ + G A + KRS +++
Sbjct: 409 EGRGVEGKTKKASKGQNSVAEKSPV--------KVARSGMADALAEMTKRSSYFQQIEED 460
Query: 57 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 116
V+ Y I ++ +H K + +S + I EKL E+ LAR+ P
Sbjct: 461 VQKYAKSIEELKSSIHSFQTKDMKELLEFHSKVESILEKLTDETQVLARFEGFPEKKLEV 520
Query: 117 IQTA 120
I+TA
Sbjct: 521 IRTA 524
>At3g07600 unknown protein
Length = 157
Score = 33.9 bits (76), Expect = 0.032
Identities = 17/29 (58%), Positives = 20/29 (68%), Gaps = 2/29 (6%)
Query: 5 GEKKPAEKKPAEEKKSTVAEKAPAEKKPK 33
G+KKP E+K EEKK EK P EKKP+
Sbjct: 80 GDKKPEEEKKPEEKKP--EEKKPEEKKPE 106
Score = 31.6 bits (70), Expect = 0.16
Identities = 17/32 (53%), Positives = 19/32 (59%), Gaps = 2/32 (6%)
Query: 8 KPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLP 39
K +KKP EEKK EK P EKKP+ K P
Sbjct: 78 KDGDKKPEEEKKPE--EKKPEEKKPEEKKPEP 107
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 33.5 bits (75), Expect = 0.042
Identities = 20/52 (38%), Positives = 28/52 (53%), Gaps = 3/52 (5%)
Query: 7 KKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVE 58
++PA P EEK + AP E AG+K +E +AA KKK+ +K E
Sbjct: 317 ERPASSTPVEEKAAQPEPVAPVEN---AGEKEGEEETAAAKKKKKKKEKEKE 365
>At3g28770 hypothetical protein
Length = 2081
Score = 33.1 bits (74), Expect = 0.055
Identities = 18/47 (38%), Positives = 24/47 (50%)
Query: 8 KPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSK 54
K EKK EEKKS E+A EKK KK ++ KK++ +
Sbjct: 1008 KNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEE 1054
Score = 28.9 bits (63), Expect = 1.0
Identities = 16/53 (30%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKN 56
+ E+K +K + KK T E+ +K+ K K PK+ DKK +K++
Sbjct: 1199 ESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKD------DKKNTTKQS 1245
Score = 28.5 bits (62), Expect = 1.3
Identities = 17/52 (32%), Positives = 25/52 (47%)
Query: 7 KKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVE 58
K EKK +E+ S EK E+K K+ K+ + DKK+ K + E
Sbjct: 994 KDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEE 1045
Score = 28.5 bits (62), Expect = 1.3
Identities = 19/60 (31%), Positives = 29/60 (47%), Gaps = 2/60 (3%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAG--DKKKRSKKNVETYK 61
K + K E+K +EE+KS ++ + K K ++ KE + KKK KK E K
Sbjct: 1032 KSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNK 1091
Score = 27.3 bits (59), Expect = 3.0
Identities = 17/55 (30%), Positives = 24/55 (42%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVE 58
K E EKK EEK T ++ +K + KK K K+K K++ E
Sbjct: 1148 KKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEE 1202
Score = 26.9 bits (58), Expect = 3.9
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query: 2 APKGEKKPAEKKPAEEKKSTVAEKAPAEK-----KPKAGKKLPKEGGSAAGDKKKRSKKN 56
A K E++ EKK +E KS E + K + KK K+ + KK+ KK+
Sbjct: 1060 AKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKD 1119
Query: 57 VE 58
+E
Sbjct: 1120 ME 1121
Score = 26.6 bits (57), Expect = 5.1
Identities = 19/52 (36%), Positives = 26/52 (49%), Gaps = 5/52 (9%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKK 55
K EKK K EE K T+ +++K K KK KE ++ KK+ KK
Sbjct: 918 KDEKKEGNK---EENKDTI--NTSSKQKGKDKKKKKKESKNSNMKKKEEDKK 964
Score = 26.2 bits (56), Expect = 6.7
Identities = 16/58 (27%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVETYK 61
K E K +KK ++K+ EK E+K K K+ ++ + +++ + KK E +K
Sbjct: 1023 KEEAKKEKKKSQDKKRE---EKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHK 1077
>At5g61610 putative protein
Length = 225
Score = 32.7 bits (73), Expect = 0.071
Identities = 22/53 (41%), Positives = 26/53 (48%), Gaps = 7/53 (13%)
Query: 4 KGEKKPAEKK-PAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKK 55
KG+K E K PAEE EK P + KP G K P++ A GDK K
Sbjct: 143 KGDKPVEEDKLPAEE------EKPPQKDKPAEGHKPPQKDKPAEGDKPVEEDK 189
Score = 32.0 bits (71), Expect = 0.12
Identities = 19/54 (35%), Positives = 26/54 (47%), Gaps = 1/54 (1%)
Query: 6 EKKPAEK-KPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVE 58
+K PAE+ KP ++ K K P + KP G K +E DK K+VE
Sbjct: 151 DKLPAEEEKPPQKDKPAEGHKPPQKDKPAEGDKPVEEDKPPQKDKPAEGDKHVE 204
Score = 31.2 bits (69), Expect = 0.21
Identities = 15/38 (39%), Positives = 20/38 (52%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKE 41
+G K P + KPAE K +K P + KP G K +E
Sbjct: 168 EGHKPPQKDKPAEGDKPVEEDKPPQKDKPAEGDKHVEE 205
Score = 26.2 bits (56), Expect = 6.7
Identities = 16/59 (27%), Positives = 25/59 (42%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVETYKI 62
+G + K AE K + + E KP KLP++ + D + K VE K+
Sbjct: 95 RGGRPDGPNKLAESGKQSGGDNPLKEDKPPERDKLPRKDKPSKEDNLLKGDKPVEEDKL 153
>At3g05060 putative SAR DNA-binding protein-1
Length = 533
Score = 32.7 bits (73), Expect = 0.071
Identities = 21/56 (37%), Positives = 31/56 (54%), Gaps = 2/56 (3%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKKNVET 59
K +KK E+KP EE+ S +K AE + +A ++ KE +KKKR + ET
Sbjct: 461 KKKKKVEEEKPEEEEPSEKKKKKKAEAETEAVVEVAKE--EKKKNKKKRKHEEEET 514
Score = 32.3 bits (72), Expect = 0.093
Identities = 20/58 (34%), Positives = 29/58 (49%), Gaps = 8/58 (13%)
Query: 6 EKKPAEKKPAEEKKS--------TVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKRSKK 55
E+KP E++P+E+KK V E A EKK K+ +E + KK+ KK
Sbjct: 468 EEKPEEEEPSEKKKKKKAEAETEAVVEVAKEEKKKNKKKRKHEEEETTETPAKKKDKK 525
Score = 29.3 bits (64), Expect = 0.79
Identities = 18/55 (32%), Positives = 31/55 (55%), Gaps = 7/55 (12%)
Query: 7 KKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGG------SAAGDKKKRSKK 55
++P++KK ++KK V E+ P E++P KK K A ++KK++KK
Sbjct: 452 EEPSKKKDKKKKKK-VEEEKPEEEEPSEKKKKKKAEAETEAVVEVAKEEKKKNKK 505
Score = 26.2 bits (56), Expect = 6.7
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKK 37
K EKK +KK E++ T + PA+KK K KK
Sbjct: 497 KEEKKKNKKKRKHEEEETT--ETPAKKKDKKEKK 528
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.306 0.125 0.328
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,071,242
Number of Sequences: 26719
Number of extensions: 125936
Number of successful extensions: 1233
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 766
Number of HSP's gapped (non-prelim): 403
length of query: 147
length of database: 11,318,596
effective HSP length: 90
effective length of query: 57
effective length of database: 8,913,886
effective search space: 508091502
effective search space used: 508091502
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC139841.15


