

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.13 - phase: 0
(148 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
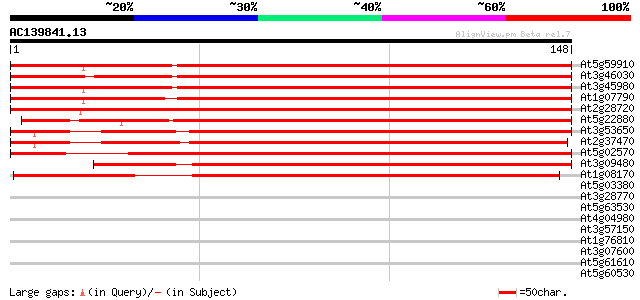
Score E
Sequences producing significant alignments: (bits) Value
At5g59910 histone H2B - like protein 254 2e-68
At3g46030 histone H2B -like protein 254 2e-68
At3g45980 histone H2B 251 8e-68
At1g07790 histone H2B 248 8e-67
At2g28720 putative histone H2B 246 2e-66
At5g22880 histone H2B like protein (emb|CAA69025.1) 238 7e-64
At3g53650 histone H2B - like protein 231 1e-61
At2g37470 putative histone H2B 223 2e-59
At5g02570 putative protein 218 9e-58
At3g09480 putative histone H2B 204 2e-53
At1g08170 hypothetical protein 132 5e-32
At5g03380 farnesylated protein - like 38 0.002
At3g28770 hypothetical protein 37 0.004
At5g63530 putative metal-binding protein 36 0.007
At4g04980 35 0.019
At3g57150 putative pseudouridine synthase (NAP57) 35 0.019
At1g76810 translation initiation factor IF-2 like protein 34 0.025
At3g07600 unknown protein 34 0.033
At5g61610 putative protein 33 0.043
At5g60530 late embryonic abundant protein - like 33 0.056
>At5g59910 histone H2B - like protein
Length = 150
Score = 254 bits (648), Expect = 2e-68
Identities = 134/151 (88%), Positives = 139/151 (91%), Gaps = 4/151 (2%)
Query: 1 MAPKGEKKPAEKKPAEEK---KSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKN 57
MAPK EKKPAEKKPA EK + + AEKAPAEKKPKAGKKLPKE G A GDKKKK KK+
Sbjct: 1 MAPKAEKKPAEKKPASEKPVEEKSKAEKAPAEKKPKAGKKLPKEAG-AGGDKKKKMKKKS 59
Query: 58 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 117
VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQE+S+LARYNKKPTITSRE
Sbjct: 60 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQEASKLARYNKKPTITSRE 119
Query: 118 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 120 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 150
>At3g46030 histone H2B -like protein
Length = 145
Score = 254 bits (648), Expect = 2e-68
Identities = 134/148 (90%), Positives = 136/148 (91%), Gaps = 3/148 (2%)
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
MAPK EKKPAEKKP EEK AEKAPAEKKPKAGKKLPKE G A GDKKKK KK+VET
Sbjct: 1 MAPKAEKKPAEKKPVEEKSK--AEKAPAEKKPKAGKKLPKEAG-AGGDKKKKMKKKSVET 57
Query: 61 YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 120
YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLA ESS+LARYNKKPTITSREIQT
Sbjct: 58 YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLASESSKLARYNKKPTITSREIQT 117
Query: 121 AVRLVLPGELAKHAVSEGTKAVTKFTSS 148
AVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 118 AVRLVLPGELAKHAVSEGTKAVTKFTSS 145
>At3g45980 histone H2B
Length = 150
Score = 251 bits (642), Expect = 8e-68
Identities = 133/151 (88%), Positives = 138/151 (91%), Gaps = 4/151 (2%)
Query: 1 MAPKGEKKPAEKKPAEEK---KSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKN 57
MAP+ EKKPAEKKPA EK + + AEKAPAEKKPKAGKKLPKE G A GDKKKK KK+
Sbjct: 1 MAPRAEKKPAEKKPAAEKPVEEKSKAEKAPAEKKPKAGKKLPKEAG-AGGDKKKKMKKKS 59
Query: 58 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 117
VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLA ESS+LARYNKKPTITSRE
Sbjct: 60 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLASESSKLARYNKKPTITSRE 119
Query: 118 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 120 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 150
>At1g07790 histone H2B
Length = 148
Score = 248 bits (633), Expect = 8e-67
Identities = 132/151 (87%), Positives = 138/151 (90%), Gaps = 6/151 (3%)
Query: 1 MAPKGEKKPAEKKPAEEK---KSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKN 57
MAP+ EKKPAEKK A E+ ++ AEKAPAEKKPKAGKKLP + AGDKKKKRSKKN
Sbjct: 1 MAPRAEKKPAEKKTAAERPVEENKAAEKAPAEKKPKAGKKLPPK---EAGDKKKKRSKKN 57
Query: 58 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 117
VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESS+LARYNKKPTITSRE
Sbjct: 58 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSKLARYNKKPTITSRE 117
Query: 118 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 118 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
>At2g28720 putative histone H2B
Length = 151
Score = 246 bits (629), Expect = 2e-66
Identities = 129/151 (85%), Positives = 137/151 (90%), Gaps = 3/151 (1%)
Query: 1 MAPKGEKKPAEKKPAEE---KKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKN 57
MAPK KKPAEKKPAE+ ++ VAEKAPAEKKPKAGKKLPKE + +KKKKR KK+
Sbjct: 1 MAPKAGKKPAEKKPAEKAPAEEEKVAEKAPAEKKPKAGKKLPKEAVTGGVEKKKKRVKKS 60
Query: 58 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 117
ETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQE+S+LARYNKKPTITSRE
Sbjct: 61 TETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQEASKLARYNKKPTITSRE 120
Query: 118 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 121 IQTAVRLVLPGELAKHAVSEGTKAVTKFTSS 151
>At5g22880 histone H2B like protein (emb|CAA69025.1)
Length = 145
Score = 238 bits (608), Expect = 7e-64
Identities = 130/146 (89%), Positives = 134/146 (91%), Gaps = 4/146 (2%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPA-EKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYK 62
K +KKPAEKKPAE K+ AE A A EKKPKAGKKLPKE + AGDKKKKRSKKNVETYK
Sbjct: 3 KADKKPAEKKPAE--KTPAAEPAAAAEKKPKAGKKLPKEP-AGAGDKKKKRSKKNVETYK 59
Query: 63 IYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAV 122
IYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLA ESS+LARYNKKPTITSREIQTAV
Sbjct: 60 IYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAGESSKLARYNKKPTITSREIQTAV 119
Query: 123 RLVLPGELAKHAVSEGTKAVTKFTSS 148
RLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 120 RLVLPGELAKHAVSEGTKAVTKFTSS 145
>At3g53650 histone H2B - like protein
Length = 138
Score = 231 bits (589), Expect = 1e-61
Identities = 126/149 (84%), Positives = 131/149 (87%), Gaps = 12/149 (8%)
Query: 1 MAPKG-EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
MAPK EKKPA KKPAE KAPAEK PKA KK+ KEGGS +KKKK+SKKN+E
Sbjct: 1 MAPKAAEKKPAGKKPAE--------KAPAEKLPKAEKKITKEGGS---EKKKKKSKKNIE 49
Query: 60 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 119
TYKIYIFKVLKQVHPDIGIS KAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ
Sbjct: 50 TYKIYIFKVLKQVHPDIGISGKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 109
Query: 120 TAVRLVLPGELAKHAVSEGTKAVTKFTSS 148
TAVRLVLPGEL+KHAVSEGTKAVTKFTSS
Sbjct: 110 TAVRLVLPGELSKHAVSEGTKAVTKFTSS 138
>At2g37470 putative histone H2B
Length = 138
Score = 223 bits (569), Expect = 2e-59
Identities = 121/148 (81%), Positives = 129/148 (86%), Gaps = 11/148 (7%)
Query: 1 MAPKG-EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
MAPK EKKPAEKKPA KAPAEK PKA KK+ K+ G + +KKKK+SKK+VE
Sbjct: 1 MAPKAAEKKPAEKKPAG--------KAPAEKLPKAEKKISKDAGGS--EKKKKKSKKSVE 50
Query: 60 TYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQ 119
TYKIYIFKVLKQVHPD+GIS KAMGIMNSFINDIFEKLAQESS+LARYNKKPTITSREIQ
Sbjct: 51 TYKIYIFKVLKQVHPDVGISGKAMGIMNSFINDIFEKLAQESSKLARYNKKPTITSREIQ 110
Query: 120 TAVRLVLPGELAKHAVSEGTKAVTKFTS 147
TAVRLVLPGELAKHAVSEGTKAVTKFTS
Sbjct: 111 TAVRLVLPGELAKHAVSEGTKAVTKFTS 138
>At5g02570 putative protein
Length = 132
Score = 218 bits (555), Expect = 9e-58
Identities = 117/148 (79%), Positives = 123/148 (83%), Gaps = 16/148 (10%)
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
MAPK EKKPAEK PA PKA KK+ KEGG++ KKKK++KK+ ET
Sbjct: 1 MAPKAEKKPAEKAPA----------------PKAEKKIAKEGGTSEIVKKKKKTKKSTET 44
Query: 61 YKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 120
YKIYIFKVLKQVHPDIGIS KAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT
Sbjct: 45 YKIYIFKVLKQVHPDIGISGKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQT 104
Query: 121 AVRLVLPGELAKHAVSEGTKAVTKFTSS 148
AVRLVLPGELAKHAVSEGTKAVTKFTSS
Sbjct: 105 AVRLVLPGELAKHAVSEGTKAVTKFTSS 132
>At3g09480 putative histone H2B
Length = 126
Score = 204 bits (518), Expect = 2e-53
Identities = 105/126 (83%), Positives = 114/126 (90%), Gaps = 4/126 (3%)
Query: 23 AEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKIYIFKVLKQVHPDIGISSKA 82
AEK P+EK PKA KK+ KEGGS ++KK++KK+ ETYKIY+FKVLKQVHPDIGIS KA
Sbjct: 5 AEKKPSEKAPKADKKITKEGGS----ERKKKTKKSTETYKIYLFKVLKQVHPDIGISGKA 60
Query: 83 MGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTAVRLVLPGELAKHAVSEGTKAV 142
MGIMNSFIND FEK+A ESSRLARYNKKPTITSREIQTAVRLVLPGELAKHAVSEGTKAV
Sbjct: 61 MGIMNSFINDTFEKIALESSRLARYNKKPTITSREIQTAVRLVLPGELAKHAVSEGTKAV 120
Query: 143 TKFTSS 148
TKFTSS
Sbjct: 121 TKFTSS 126
>At1g08170 hypothetical protein
Length = 243
Score = 132 bits (333), Expect = 5e-32
Identities = 68/144 (47%), Positives = 97/144 (67%), Gaps = 15/144 (10%)
Query: 2 APKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETY 61
+P+ + PA K KK+ EK KK K KKKKR + Y
Sbjct: 107 SPQPPETPASKSEGTLKKTDKVEKKQENKKKK---------------KKKKRDDLAGDEY 151
Query: 62 KIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSREIQTA 121
+ Y++KV+KQVHPD+GI+SKAM ++N F+ D+FE++AQE++RL+ Y K+ T++SREI+ A
Sbjct: 152 RRYVYKVMKQVHPDLGITSKAMTVVNMFMGDMFERIAQEAARLSDYTKRRTLSSREIEAA 211
Query: 122 VRLVLPGELAKHAVSEGTKAVTKF 145
VRLVLPGEL++HAV+EG+KAV+ F
Sbjct: 212 VRLVLPGELSRHAVAEGSKAVSNF 235
>At5g03380 farnesylated protein - like
Length = 392
Score = 38.1 bits (87), Expect = 0.002
Identities = 22/59 (37%), Positives = 33/59 (55%), Gaps = 8/59 (13%)
Query: 1 MAPKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
+AP ++ P AE+K S AE+ PAEKKP A +K G+KK+++ K+ E
Sbjct: 92 VAPPKKETPPSSGGAEKKPSPAAEEKPAEKKPAAVEK--------PGEKKEEKKKEEGE 142
Score = 26.6 bits (57), Expect = 5.2
Identities = 14/37 (37%), Positives = 20/37 (53%), Gaps = 6/37 (16%)
Query: 2 APKGEKKPAEKKPA------EEKKSTVAEKAPAEKKP 32
+P E+KPAEKKPA E+K+ E+ + P
Sbjct: 111 SPAAEEKPAEKKPAAVEKPGEKKEEKKKEEGEKKASP 147
>At3g28770 hypothetical protein
Length = 2081
Score = 37.0 bits (84), Expect = 0.004
Identities = 20/55 (36%), Positives = 28/55 (50%)
Query: 8 KPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYK 62
K EKK EEKKS E+A EKK KK ++ KK+K ++++ K
Sbjct: 1008 KNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKK 1062
Score = 30.4 bits (67), Expect = 0.36
Identities = 20/60 (33%), Positives = 30/60 (49%), Gaps = 1/60 (1%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDK-KKKRSKKNVETYK 62
K + K E+K +EE+KS ++ + K K ++ KE + K KKK KK E K
Sbjct: 1032 KSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNK 1091
Score = 28.1 bits (61), Expect = 1.8
Identities = 16/51 (31%), Positives = 26/51 (50%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKK 56
+KK + ++KK K EKK K ++ K+ S +KK+K +KK
Sbjct: 1179 DKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKK 1229
Score = 28.1 bits (61), Expect = 1.8
Identities = 20/60 (33%), Positives = 27/60 (44%), Gaps = 2/60 (3%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
K E EKK EEK T ++ +K + KK K S KKK++ K E K+
Sbjct: 1148 KKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKK--SSKDQQKKKEKEMKESEEKKL 1205
Score = 27.7 bits (60), Expect = 2.3
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEK---KPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
K EK+ + A++K+ EK +E K K KK ++ S ++ KK KK+ E+
Sbjct: 1049 KKEKEESRDLKAKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEES 1108
Score = 27.7 bits (60), Expect = 2.3
Identities = 17/59 (28%), Positives = 29/59 (48%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYK 62
K EKK ++ +K+S EK E+K + + + DKK+K+S K+ + K
Sbjct: 1135 KNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKK 1193
Score = 27.3 bits (59), Expect = 3.1
Identities = 16/54 (29%), Positives = 26/54 (47%), Gaps = 7/54 (12%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKN 57
+ E+K +K + KK T E+ +K+ K K PK D KK +K++
Sbjct: 1199 ESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPK-------DDKKNTTKQS 1245
Score = 27.3 bits (59), Expect = 3.1
Identities = 19/62 (30%), Positives = 30/62 (47%), Gaps = 4/62 (6%)
Query: 2 APKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKK----LPKEGGSAAGDKKKKRSKKN 57
A K E++ EKK +E KS E + K+ KK K+ + +KK+ KK+
Sbjct: 1060 AKKKEEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKD 1119
Query: 58 VE 59
+E
Sbjct: 1120 ME 1121
Score = 26.2 bits (56), Expect = 6.8
Identities = 16/60 (26%), Positives = 28/60 (46%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
K KK +KK E+ + + K +K K + K + K+KK +++ ET +I
Sbjct: 1109 KSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEI 1168
Score = 26.2 bits (56), Expect = 6.8
Identities = 16/57 (28%), Positives = 29/57 (50%), Gaps = 2/57 (3%)
Query: 6 EKKPAEKKPA--EEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
EKK K+ A E+KKS ++ + + + KK +E KK++ +K+ E+
Sbjct: 1017 EKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKES 1073
Score = 26.2 bits (56), Expect = 6.8
Identities = 15/55 (27%), Positives = 24/55 (43%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
E K +K+ E KK+ E K+ + G+KK+ + K+VET
Sbjct: 747 EAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVET 801
Score = 26.2 bits (56), Expect = 6.8
Identities = 18/53 (33%), Positives = 25/53 (46%), Gaps = 6/53 (11%)
Query: 7 KKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
K EKK +E+ S EK E+K K+ K+ +KKK + KK E
Sbjct: 994 KDNKEKKESEDSASKNREKKEYEEKKSKTKEEAKK------EKKKSQDKKREE 1040
Score = 25.8 bits (55), Expect = 8.9
Identities = 23/68 (33%), Positives = 29/68 (41%), Gaps = 12/68 (17%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVETYKI 63
K EKK K EE K T+ +++K K KK KE K KK E K
Sbjct: 918 KDEKKEGNK---EENKDTI--NTSSKQKGKDKKKKKKE-------SKNSNMKKKEEDKKE 965
Query: 64 YIFKVLKQ 71
Y+ LK+
Sbjct: 966 YVNNELKK 973
>At5g63530 putative metal-binding protein
Length = 355
Score = 36.2 bits (82), Expect = 0.007
Identities = 22/45 (48%), Positives = 26/45 (56%), Gaps = 1/45 (2%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKK 50
EKK EKKP EEKK +K AEKK + K P+EG S K+
Sbjct: 13 EKKMEEKKP-EEKKEGEDKKVDAEKKGEDSDKKPQEGESNKDSKE 56
Score = 33.9 bits (76), Expect = 0.033
Identities = 17/50 (34%), Positives = 25/50 (50%)
Query: 11 EKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVET 60
EKKP ++ + EK P EKK KK+ E DKK + + N ++
Sbjct: 5 EKKPEAAEEKKMEEKKPEEKKEGEDKKVDAEKKGEDSDKKPQEGESNKDS 54
>At4g04980
Length = 681
Score = 34.7 bits (78), Expect = 0.019
Identities = 32/124 (25%), Positives = 56/124 (44%), Gaps = 14/124 (11%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRS------KKN 57
+G + K A + +++VAEK+P K+ + G + A + KRS +++
Sbjct: 409 EGRGVEGKTKKASKGQNSVAEKSPV--------KVARSGMADALAEMTKRSSYFQQIEED 460
Query: 58 VETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSRLARYNKKPTITSRE 117
V+ Y I ++ +H K + +S + I EKL E+ LAR+ P
Sbjct: 461 VQKYAKSIEELKSSIHSFQTKDMKELLEFHSKVESILEKLTDETQVLARFEGFPEKKLEV 520
Query: 118 IQTA 121
I+TA
Sbjct: 521 IRTA 524
>At3g57150 putative pseudouridine synthase (NAP57)
Length = 565
Score = 34.7 bits (78), Expect = 0.019
Identities = 21/57 (36%), Positives = 31/57 (53%), Gaps = 5/57 (8%)
Query: 8 KPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEG-----GSAAGDKKKKRSKKNVE 59
K ++KK ++K+ E+A +EKK K KK KE S +KKKK+ K+ E
Sbjct: 474 KSSKKKKKKDKEEEKEEEAGSEKKEKKKKKDKKEEVIEEVASPKSEKKKKKKSKDTE 530
Score = 31.6 bits (70), Expect = 0.16
Identities = 21/53 (39%), Positives = 30/53 (55%), Gaps = 4/53 (7%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKK 56
K + K E + AEEK + +K +KK K +K +E GS +KKKK+ KK
Sbjct: 458 KSKTKEVEGEEAEEKVKSSKKK---KKKDKEEEK-EEEAGSEKKEKKKKKDKK 506
Score = 31.2 bits (69), Expect = 0.21
Identities = 23/60 (38%), Positives = 30/60 (49%), Gaps = 10/60 (16%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPK--------EGGSAAGDKKKKRSKKN 57
EKK EKK ++KK V E+ + K K KK K E SAA +KK+ KK+
Sbjct: 495 EKK--EKKKKKDKKEEVIEEVASPKSEKKKKKKSKDTEAAVDAEDESAAEKSEKKKKKKD 552
Score = 28.1 bits (61), Expect = 1.8
Identities = 19/58 (32%), Positives = 27/58 (45%), Gaps = 7/58 (12%)
Query: 2 APKGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKKNVE 59
+PK EKK +K E +++ AEK K KK K+ KK K S+ + E
Sbjct: 515 SPKSEKKKKKKSKDTEAAVDAEDESAAEKSEKKKKKKDKK-------KKNKDSEDDEE 565
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 34.3 bits (77), Expect = 0.025
Identities = 20/50 (40%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query: 7 KKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKK 56
++PA P EEK + AP E AG+K +E +AA KKKK+ K+
Sbjct: 317 ERPASSTPVEEKAAQPEPVAPVEN---AGEKEGEEETAAAKKKKKKKEKE 363
>At3g07600 unknown protein
Length = 157
Score = 33.9 bits (76), Expect = 0.033
Identities = 17/29 (58%), Positives = 20/29 (68%), Gaps = 2/29 (6%)
Query: 5 GEKKPAEKKPAEEKKSTVAEKAPAEKKPK 33
G+KKP E+K EEKK EK P EKKP+
Sbjct: 80 GDKKPEEEKKPEEKKP--EEKKPEEKKPE 106
Score = 31.6 bits (70), Expect = 0.16
Identities = 17/32 (53%), Positives = 19/32 (59%), Gaps = 2/32 (6%)
Query: 8 KPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLP 39
K +KKP EEKK EK P EKKP+ K P
Sbjct: 78 KDGDKKPEEEKKPE--EKKPEEKKPEEKKPEP 107
>At5g61610 putative protein
Length = 225
Score = 33.5 bits (75), Expect = 0.043
Identities = 22/53 (41%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query: 4 KGEKKPAEKK-PAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSK 55
KG+K E K PAEE EK P + KP G K P++ A GDK + K
Sbjct: 143 KGDKPVEEDKLPAEE------EKPPQKDKPAEGHKPPQKDKPAEGDKPVEEDK 189
Score = 31.2 bits (69), Expect = 0.21
Identities = 15/38 (39%), Positives = 20/38 (52%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKE 41
+G K P + KPAE K +K P + KP G K +E
Sbjct: 168 EGHKPPQKDKPAEGDKPVEEDKPPQKDKPAEGDKHVEE 205
Score = 30.4 bits (67), Expect = 0.36
Identities = 17/44 (38%), Positives = 21/44 (47%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDK 49
EK P + KPAE K +K KP K P++ A GDK
Sbjct: 158 EKPPQKDKPAEGHKPPQKDKPAEGDKPVEEDKPPQKDKPAEGDK 201
Score = 28.1 bits (61), Expect = 1.8
Identities = 17/51 (33%), Positives = 24/51 (46%), Gaps = 1/51 (1%)
Query: 6 EKKPAEK-KPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSK 55
+K PAE+ KP ++ K K P + KP G K +E DK + K
Sbjct: 151 DKLPAEEEKPPQKDKPAEGHKPPQKDKPAEGDKPVEEDKPPQKDKPAEGDK 201
>At5g60530 late embryonic abundant protein - like
Length = 439
Score = 33.1 bits (74), Expect = 0.056
Identities = 22/61 (36%), Positives = 29/61 (47%), Gaps = 1/61 (1%)
Query: 4 KGEKKPAEKKPAEEKKSTVAEKAPAEKKPKAGK-KLPKEGGSAAGDKKKKRSKKNVETYK 62
K +K+ EKK E K+ +K EKK K K K KE A +KK K + Y+
Sbjct: 80 KKDKEEKEKKDKERKEKEKKDKLEKEKKDKERKEKERKEKERKAKEKKDKEESEAAARYR 139
Query: 63 I 63
I
Sbjct: 140 I 140
Score = 29.6 bits (65), Expect = 0.62
Identities = 17/51 (33%), Positives = 25/51 (48%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRSKK 56
EKK EK ++K+ +K EKK K K+ K+ K K+R +K
Sbjct: 64 EKKDKEKAAKDKKEKEKKDKEEKEKKDKERKEKEKKDKLEKEKKDKERKEK 114
Score = 29.3 bits (64), Expect = 0.80
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 3/57 (5%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGKKLPKEGGSAAGDKKKKRS---KKNVE 59
+KK EKK EEK+ E+ EKK K K+ + K+K+R KK+ E
Sbjct: 74 DKKEKEKKDKEEKEKKDKERKEKEKKDKLEKEKKDKERKEKERKEKERKAKEKKDKE 130
Score = 28.5 bits (62), Expect = 1.4
Identities = 18/52 (34%), Positives = 26/52 (49%), Gaps = 1/52 (1%)
Query: 6 EKKPAEKKPAEEKKSTVAEKAPAEKKPKAGK-KLPKEGGSAAGDKKKKRSKK 56
EK +KK E+K EK E+K K K KL KE +K+++ K+
Sbjct: 69 EKAAKDKKEKEKKDKEEKEKKDKERKEKEKKDKLEKEKKDKERKEKERKEKE 120
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.306 0.125 0.327
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,112,047
Number of Sequences: 26719
Number of extensions: 129820
Number of successful extensions: 1403
Number of sequences better than 10.0: 170
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 840
Number of HSP's gapped (non-prelim): 479
length of query: 148
length of database: 11,318,596
effective HSP length: 90
effective length of query: 58
effective length of database: 8,913,886
effective search space: 517005388
effective search space used: 517005388
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC139841.13


