

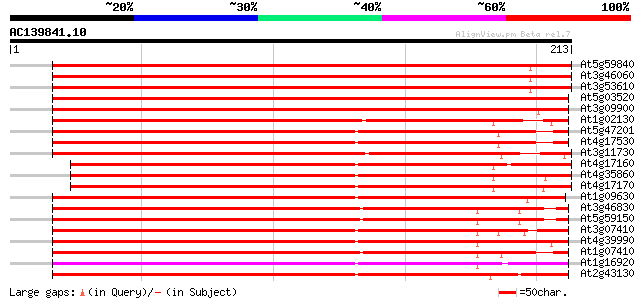
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.10 - phase: 0
(213 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59840 GTP-binding protein ara-3 358 e-100
At3g46060 GTP-binding protein ara-3 358 1e-99
At3g53610 GTPase AtRAB8 352 8e-98
At5g03520 GTP-binding protein - like 333 3e-92
At3g09900 putative Ras-like GTP-binding protein 324 2e-89
At1g02130 GTP-binding protein, ara-5 232 1e-61
At5g47201 ras-related small GTP-binding protein-like 214 2e-56
At4g17530 ras-related small GTP-binding protein RAB1c 214 3e-56
At3g11730 putative GTP-binding protein (ATFP8) 211 2e-55
At4g17160 GTP-binding RAB2A like protein 174 2e-44
At4g35860 GTP-binding protein GB2 174 4e-44
At4g17170 GTP-binding RAB2A like protein 171 2e-43
At1g09630 putative RAS-related protein, RAB11C 161 2e-40
At3g46830 GTP-binding protein Rab11 159 8e-40
At5g59150 GTP-binding protein rab11 - like 159 1e-39
At3g07410 GTP-binding protein like 158 2e-39
At4g39990 GTP-binding protein GB3 157 4e-39
At1g07410 small G protein, putative 156 9e-39
At1g16920 unknown protein 155 1e-38
At2g43130 Ras-related GTP-binding protein (ARA-4) 152 1e-37
>At5g59840 GTP-binding protein ara-3
Length = 216
Score = 358 bits (920), Expect = e-100
Identities = 181/198 (91%), Positives = 190/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVP SKGQALADEYGIKFFETSAKTNLNVEEVFFSIA+DIKQRLADTD++AEP TIKI+Q
Sbjct: 138 AVPKSKGQALADEYGIKFFETSAKTNLNVEEVFFSIAKDIKQRLADTDSRAEPATIKISQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A G+GQA QKSACCG
Sbjct: 198 TDQAAGAGQATQKSACCG 215
>At3g46060 GTP-binding protein ara-3
Length = 216
Score = 358 bits (919), Expect = 1e-99
Identities = 180/198 (90%), Positives = 190/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPT+KGQALADEYGIKFFETSAKTNLNVEEVFFSI RDIKQRL+DTD++AEP TIKI+Q
Sbjct: 138 AVPTAKGQALADEYGIKFFETSAKTNLNVEEVFFSIGRDIKQRLSDTDSRAEPATIKISQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A G+GQA QKSACCG
Sbjct: 198 TDQAAGAGQATQKSACCG 215
>At3g53610 GTPase AtRAB8
Length = 216
Score = 352 bits (903), Expect = 8e-98
Identities = 178/198 (89%), Positives = 187/198 (93%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASD+VNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDSVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVP SKGQALADEYG+KFFETSAKTNLNVEEVFFSIA+DIKQRLADTD +AEP TIKINQ
Sbjct: 138 AVPKSKGQALADEYGMKFFETSAKTNLNVEEVFFSIAKDIKQRLADTDARAEPQTIKINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D G+ QA QKSACCG
Sbjct: 198 SDQGAGTSQATQKSACCG 215
>At5g03520 GTP-binding protein - like
Length = 216
Score = 333 bits (855), Expect = 3e-92
Identities = 165/196 (84%), Positives = 178/196 (90%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSD +FTTSFITTIGIDFKIRT+ELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDDTFTTSFITTIGIDFKIRTVELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNW++NIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWMKNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPT+KGQALADEYGIKFFETSAKTNLNVE VF SIA+DIKQRL +TD KAEP IKI +
Sbjct: 138 AVPTAKGQALADEYGIKFFETSAKTNLNVENVFMSIAKDIKQRLTETDTKAEPQGIKITK 197
Query: 197 DSATGSGQAAQKSACC 212
S A+KSACC
Sbjct: 198 QDTAASSSTAEKSACC 213
>At3g09900 putative Ras-like GTP-binding protein
Length = 218
Score = 324 bits (830), Expect = 2e-89
Identities = 163/198 (82%), Positives = 178/198 (89%), Gaps = 2/198 (1%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSD +FTTSFITTIGIDFKIRT+ELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDDTFTTSFITTIGIDFKIRTVELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNW++NIEQHASD+VNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWMKNIEQHASDSVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTN NVE+VF SIA+DIKQRL ++D KAEP IKI +
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNQNVEQVFLSIAKDIKQRLTESDTKAEPQGIKITK 197
Query: 197 DSA--TGSGQAAQKSACC 212
A S +KSACC
Sbjct: 198 QDANKASSSSTNEKSACC 215
>At1g02130 GTP-binding protein, ara-5
Length = 203
Score = 232 bits (592), Expect = 1e-61
Identities = 118/199 (59%), Positives = 151/199 (75%), Gaps = 11/199 (5%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSD S+ S+I+TIG+DFKIRT+E DGK IKLQIWDTAGQERF
Sbjct: 11 LLLIGDSGVGKSCLLLRFSDDSYVESYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQERF 70
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTIT++YYRGA GI++VYDVTDE SFNN++ W+ I+++ASDNVNK+LVGNK+D+ E+ R
Sbjct: 71 RTITSSYYRGAHGIIIVYDVTDEESFNNVKQWLSEIDRYASDNVNKLLVGNKSDLTEN-R 129
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDNKAEPTTIKI 194
A+P +A ADE GI F ETSAK NVE+ F +++ IK+R+A N A P T++I
Sbjct: 130 AIPYETAKAFADEIGIPFMETSAKDATNVEQAFMAMSASIKERMASQPAGNNARPPTVQI 189
Query: 195 NQDSATGSGQ-AAQKSACC 212
GQ AQK+ CC
Sbjct: 190 R-------GQPVAQKNGCC 201
>At5g47201 ras-related small GTP-binding protein-like
Length = 202
Score = 214 bits (546), Expect = 2e-56
Identities = 110/197 (55%), Positives = 142/197 (71%), Gaps = 8/197 (4%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRF+D S+ S+I+TIG+DFKIRT+E DGK IKLQIWDTAGQERF
Sbjct: 11 LLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQERF 70
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTIT++YYRGA GI++ YDVTD SFNN++ W+ I+++AS+NVNK+LVGNK D+ S++
Sbjct: 71 RTITSSYYRGAHGIIVTYDVTDLESFNNVKQWLNEIDRYASENVNKLLVGNKNDL-TSQK 129
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTD-NKAEPTTIKIN 195
V T +A ADE GI F ETSAK NVEE F ++ IK R+A A+P T++I
Sbjct: 130 VVSTETAKAFADELGIPFLETSAKNATNVEEAFMAMTAAIKTRMASQPAGGAKPPTVQIR 189
Query: 196 QDSATGSGQAAQKSACC 212
Q+S CC
Sbjct: 190 GQPVN------QQSGCC 200
>At4g17530 ras-related small GTP-binding protein RAB1c
Length = 202
Score = 214 bits (545), Expect = 3e-56
Identities = 109/197 (55%), Positives = 142/197 (71%), Gaps = 8/197 (4%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRF+D S+ S+I+TIG+DFKIRT+E DGK IKLQIWDTAGQERF
Sbjct: 11 LLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQERF 70
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTIT++YYRGA GI++ YDVTD SFNN++ W+ I+++AS+NVNK+LVGNK D+ S++
Sbjct: 71 RTITSSYYRGAHGIIVTYDVTDLESFNNVKQWLNEIDRYASENVNKLLVGNKCDL-TSQK 129
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTD-NKAEPTTIKIN 195
V T +A ADE GI F ETSAK NVEE F ++ IK R+A ++P T++I
Sbjct: 130 VVSTETAKAFADELGIPFLETSAKNATNVEEAFMAMTAAIKTRMASQPAGGSKPPTVQIR 189
Query: 196 QDSATGSGQAAQKSACC 212
Q+S CC
Sbjct: 190 GQPVN------QQSGCC 200
>At3g11730 putative GTP-binding protein (ATFP8)
Length = 205
Score = 211 bits (537), Expect = 2e-55
Identities = 109/202 (53%), Positives = 149/202 (72%), Gaps = 13/202 (6%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ + VGKSCLLLRF+D ++ S+I+TIG+DFKIRTIE DGK IKLQIWDTAGQERF
Sbjct: 11 LLLIGDSSVGKSCLLLRFADDAYIDSYISTIGVDFKIRTIEQDGKTIKLQIWDTAGQERF 70
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTIT++YYRGA GI++VYD T+ SFNN++ W+ I+++A+++V K+L+GNK DM ESK
Sbjct: 71 RTITSSYYRGAHGIIIVYDCTEMESFNNVKQWLSEIDRYANESVCKLLIGNKNDMVESK- 129
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDN---KAEPTTIK 193
V T G+ALADE GI F ETSAK ++NVE+ F +IA +IK+++ N + P T++
Sbjct: 130 VVSTETGRALADELGIPFLETSAKDSINVEQAFLTIAGEIKKKMGSQTNANKTSGPGTVQ 189
Query: 194 INQDSATGSGQAAQKS--ACCG 213
+ GQ Q++ CCG
Sbjct: 190 M-------KGQPIQQNNGGCCG 204
>At4g17160 GTP-binding RAB2A like protein
Length = 205
Score = 174 bits (442), Expect = 2e-44
Identities = 91/192 (47%), Positives = 120/192 (62%), Gaps = 4/192 (2%)
Query: 24 GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTAY 83
GVGKSCLLL+F+D F TIG++F +TI +D K IKLQIWDTAGQE FR++T +Y
Sbjct: 16 GVGKSCLLLKFTDKRFQAVHDLTIGVEFGAKTITIDNKPIKLQIWDTAGQESFRSVTRSY 75
Query: 84 YRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKRAVPTSKG 143
YRG G LLVYD+T +FN++ +W+ QHAS+N+ +L+GNK D+ E KR V T +G
Sbjct: 76 YRGRAGTLLVYDITRRETFNHLASWLEEARQHASENMTTMLIGNKCDL-EDKRTVSTEEG 134
Query: 144 QALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDNKAEPTTIKINQDSATG 201
+ A E+G+ F E SAKT NVEE F A I +R+ D D EP I
Sbjct: 135 EQFAREHGLIFMEASAKTAHNVEEAFVETAATIYKRIQDGVVDEANEP-GITPGPFGGKD 193
Query: 202 SGQAAQKSACCG 213
+ + Q+ CCG
Sbjct: 194 ASSSQQRRGCCG 205
>At4g35860 GTP-binding protein GB2
Length = 211
Score = 174 bits (440), Expect = 4e-44
Identities = 93/197 (47%), Positives = 123/197 (62%), Gaps = 8/197 (4%)
Query: 24 GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTAY 83
GVGKSCLLL+F+D F TIG++F R + +DG+ IKLQIWDTAGQE FR+IT +Y
Sbjct: 16 GVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMVTVDGRPIKLQIWDTAGQESFRSITRSY 75
Query: 84 YRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKRAVPTSKG 143
YRGA G LLVYD+T +FN++ +W+ + QHA+ N++ +L+GNK D+ KRAV +G
Sbjct: 76 YRGAAGALLVYDITRRETFNHLASWLEDARQHANPNMSIMLIGNKCDL-AHKRAVSKEEG 134
Query: 144 QALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDNKAEPTTIKINQDSATG 201
Q A E+G+ F E SA+T NVEE F A I Q + D D E + IKI G
Sbjct: 135 QQFAKEHGLLFLEASARTAQNVEEAFIETAAKILQNIQDGVFDVSNESSGIKIGYGRTQG 194
Query: 202 S-----GQAAQKSACCG 213
+ G +Q CCG
Sbjct: 195 AAGGRDGTISQGGGCCG 211
>At4g17170 GTP-binding RAB2A like protein
Length = 211
Score = 171 bits (434), Expect = 2e-43
Identities = 92/197 (46%), Positives = 121/197 (60%), Gaps = 8/197 (4%)
Query: 24 GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTAY 83
GVGKSCLLL+F+D F TIG++F R I +D K IKLQIWDTAGQE FR+IT +Y
Sbjct: 16 GVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDNKPIKLQIWDTAGQESFRSITRSY 75
Query: 84 YRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKRAVPTSKG 143
YRGA G LLVYD+T +FN++ +W+ + QHA+ N+ +L+GNK D+ +RAV T +G
Sbjct: 76 YRGAAGALLVYDITRRETFNHLASWLEDARQHANANMTIMLIGNKCDL-AHRRAVSTEEG 134
Query: 144 QALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDNKAEPTTIKINQDSATG 201
+ A E+G+ F E SAKT NVEE F A I +++ D D E IK+ G
Sbjct: 135 EQFAKEHGLIFMEASAKTAQNVEEAFIKTAATIYKKIQDGVFDVSNESYGIKVGYGGIPG 194
Query: 202 -----SGQAAQKSACCG 213
G +Q CCG
Sbjct: 195 PSGGRDGSTSQGGGCCG 211
>At1g09630 putative RAS-related protein, RAB11C
Length = 217
Score = 161 bits (408), Expect = 2e-40
Identities = 84/201 (41%), Positives = 125/201 (61%), Gaps = 7/201 (3%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
++L+ +GVGKS LL RF+ F +TIG++F RT++++G+ +K QIWDTAGQER+
Sbjct: 15 VVLIGDSGVGKSNLLSRFTRNEFCLESKSTIGVEFATRTLQVEGRTVKAQIWDTAGQERY 74
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R IT+AYYRGA+G LLVYDVT +F N+ W++ + HA N+ +L+GNK D+ + R
Sbjct: 75 RAITSAYYRGALGALLVYDVTKPTTFENVSRWLKELRDHADSNIVIMLIGNKTDL-KHLR 133
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKIN- 195
AV T Q+ A++ G+ F ETSA LNVE+ F +I ++ + ++ ++ TT N
Sbjct: 134 AVATEDAQSYAEKEGLSFIETSALEALNVEKAFQTILSEVYRIISKKSISSDQTTANANI 193
Query: 196 -----QDSATGSGQAAQKSAC 211
D A S A+K C
Sbjct: 194 KEGQTIDVAATSESNAKKPCC 214
>At3g46830 GTP-binding protein Rab11
Length = 217
Score = 159 bits (403), Expect = 8e-40
Identities = 89/206 (43%), Positives = 127/206 (61%), Gaps = 15/206 (7%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
I+L+ +GVGKS +L RF+ F +TIG++F RT +++GK IK QIWDTAGQER+
Sbjct: 15 IVLIGDSGVGKSNILSRFTRNEFCLESKSTIGVEFATRTTQVEGKTIKAQIWDTAGQERY 74
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R IT+AYYRGA+G LLVYD+T +F+N+ W+R + HA N+ ++ GNK+D++ R
Sbjct: 75 RAITSAYYRGAVGALLVYDITKRQTFDNVLRWLRELRDHADSNIVIMMAGNKSDLNH-LR 133
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRLADTDNKAEPTT 191
+V GQ+LA++ G+ F ETSA NVE+ F +I +I K+ LA + A +
Sbjct: 134 SVAEEDGQSLAEKEGLSFLETSALEATNVEKAFQTILGEIYHIISKKALAAQEAAAANSA 193
Query: 192 I-----KINQDSATGSGQAAQKSACC 212
I IN D +G K ACC
Sbjct: 194 IPGQGTTINVDDTSGGA----KRACC 215
>At5g59150 GTP-binding protein rab11 - like
Length = 217
Score = 159 bits (401), Expect = 1e-39
Identities = 86/206 (41%), Positives = 127/206 (60%), Gaps = 15/206 (7%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
I+L+ +GVGK+ +L RF+ F +TIG++F RT++++GK +K QIWDTAGQER+
Sbjct: 15 IVLIGDSGVGKTNILSRFTRNEFCLESKSTIGVEFATRTLQVEGKTVKAQIWDTAGQERY 74
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R IT+AYYRGA+G LLVYD+T +F+N+ W+R + HA N+ ++ GNKAD++ R
Sbjct: 75 RAITSAYYRGAVGALLVYDITKRQTFDNVLRWLRELRDHADSNIVIMMAGNKADLNH-LR 133
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRLADTDNKAEPTT 191
+V GQ LA+ G+ F ETSA NVE+ F ++ +I K+ LA + A +
Sbjct: 134 SVAEEDGQTLAETEGLSFLETSALEATNVEKAFQTVLAEIYHIISKKALAAQEAAAANSA 193
Query: 192 I-----KINQDSATGSGQAAQKSACC 212
I IN + +G+G K CC
Sbjct: 194 IPGQGTTINVEDTSGAG----KRGCC 215
>At3g07410 GTP-binding protein like
Length = 217
Score = 158 bits (400), Expect = 2e-39
Identities = 88/205 (42%), Positives = 132/205 (63%), Gaps = 13/205 (6%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
I+L+ + VGKS LL RFS F T+ TIG++F+ + +E++GK +K QIWDTAGQERF
Sbjct: 15 IVLIGDSAVGKSNLLSRFSRDEFDTNSKATIGVEFQTQLVEIEGKEVKAQIWDTAGQERF 74
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R +T+AYYRGA G L+VYD+T +F +++ W++ + H V ++LVGNK D+ E R
Sbjct: 75 RAVTSAYYRGAFGALIVYDITRGDTFESVKRWLQELNTHCDTAVAQMLVGNKCDL-EDIR 133
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI----KQRLADTD-NKAEPTT 191
AV +G+ALA+E G+ F ETSA NV++ F + R+I ++L ++D KAE +
Sbjct: 134 AVSVEEGKALAEEEGLFFMETSALDATNVDKAFEIVIREIFNNVSRKLLNSDAYKAELSV 193
Query: 192 IKI----NQDSATGSGQAAQKSACC 212
++ NQD GS + + +CC
Sbjct: 194 NRVSLVNNQD---GSESSWRNPSCC 215
>At4g39990 GTP-binding protein GB3
Length = 224
Score = 157 bits (397), Expect = 4e-39
Identities = 85/203 (41%), Positives = 124/203 (60%), Gaps = 8/203 (3%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
++L+ + VGKS LL RF+ F+ TIG++F+ RT+ ++ K IK QIWDTAGQER+
Sbjct: 20 VVLIGDSAVGKSQLLARFARDEFSMDSKATIGVEFQTRTLSIEQKSIKAQIWDTAGQERY 79
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R +T+AYYRGA+G +LVYD+T +F +I W+ + HA N+ IL+GNK+D+ E +R
Sbjct: 80 RAVTSAYYRGAVGAMLVYDMTKRETFEHIPRWLEELRAHADKNIVIILIGNKSDL-EDQR 138
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRLADTDNKAEPTT 191
AVPT + A++ G+ F ETSA NVE F ++ I K+ LA + P +
Sbjct: 139 AVPTEDAKEFAEKEGLFFLETSALNATNVENSFNTLMTQIYNTVNKKNLASEGDSNNPGS 198
Query: 192 IKINQDSATGSGQ--AAQKSACC 212
+ + GSGQ A+ S CC
Sbjct: 199 LAGKKILIPGSGQEIPAKTSTCC 221
>At1g07410 small G protein, putative
Length = 214
Score = 156 bits (394), Expect = 9e-39
Identities = 84/205 (40%), Positives = 126/205 (60%), Gaps = 16/205 (7%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
I+L+ +GVGKS +L RF+ F +TIG++F RT++++GK +K QIWDTAGQER+
Sbjct: 15 IVLIGDSGVGKSNILSRFTRNEFCLESKSTIGVEFATRTLQVEGKTVKAQIWDTAGQERY 74
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R IT+AYYRGA+G LLVYD+T +F N+ W+R + HA N+ ++ GNK+D++ R
Sbjct: 75 RAITSAYYRGAVGALLVYDITKRQTFENVLRWLRELRDHADSNIVIMMAGNKSDLNH-LR 133
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRLADTDN----KA 187
+V G++LA++ G+ F ETSA N+E+ F +I +I K+ LA +
Sbjct: 134 SVADEDGRSLAEKEGLSFLETSALEATNIEKAFQTILSEIYHIISKKALAAQEAAGNLPG 193
Query: 188 EPTTIKINQDSATGSGQAAQKSACC 212
+ T I I+ SAT + CC
Sbjct: 194 QGTAINISDSSAT------NRKGCC 212
>At1g16920 unknown protein
Length = 216
Score = 155 bits (393), Expect = 1e-38
Identities = 86/202 (42%), Positives = 121/202 (59%), Gaps = 9/202 (4%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
++L+ +GVGKS LL RF+ F +TIG++F RT+++DGK +K QIWDTAGQER+
Sbjct: 16 VVLIGDSGVGKSNLLSRFTKNEFNLESKSTIGVEFATRTLKVDGKVVKAQIWDTAGQERY 75
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R IT+AYYRGA+G LLVYDVT A+F N+ W++ ++ H N+ +LVGNK+D+
Sbjct: 76 RAITSAYYRGAVGALLVYDVTRRATFENVDRWLKELKNHTDPNIVVMLVGNKSDL-RHLL 134
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI------KQRLADTDNKAEPT 190
AVPT G++ A++ + F ETSA NVE+ F + I KQ A D A +
Sbjct: 135 AVPTEDGKSYAEQESLCFMETSALEATNVEDAFAEVLTQIYRITSKKQVEAGEDGNA--S 192
Query: 191 TIKINQDSATGSGQAAQKSACC 212
K + A +K CC
Sbjct: 193 VPKGEKIEVKNDVSALKKLGCC 214
>At2g43130 Ras-related GTP-binding protein (ARA-4)
Length = 214
Score = 152 bits (384), Expect = 1e-37
Identities = 77/200 (38%), Positives = 124/200 (61%), Gaps = 6/200 (3%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
I+++ + VGKS LL R++ F + TIG++F+ +++ +DGK +K QIWDTAGQERF
Sbjct: 15 IVIIGDSAVGKSNLLTRYARNEFNPNSKATIGVEFQTQSMLIDGKEVKAQIWDTAGQERF 74
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
R +T+AYYRGA+G L+VYD+T ++F N+ W+ + H+ V K+L+GNK D+ ES R
Sbjct: 75 RAVTSAYYRGAVGALVVYDITRSSTFENVGRWLDELNTHSDTTVAKMLIGNKCDL-ESIR 133
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLA----DTDNKAEPTTI 192
AV +G++LA+ G+ F ETSA + NV+ F + R+I ++ ++D+ E T+
Sbjct: 134 AVSVEEGKSLAESEGLFFMETSALDSTNVKTAFEMVIREIYSNISRKQLNSDSYKEELTV 193
Query: 193 KINQDSATGSGQAAQKSACC 212
+ + +CC
Sbjct: 194 N-RVSLVKNENEGTKTFSCC 212
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,316,678
Number of Sequences: 26719
Number of extensions: 163522
Number of successful extensions: 652
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 448
Number of HSP's gapped (non-prelim): 109
length of query: 213
length of database: 11,318,596
effective HSP length: 95
effective length of query: 118
effective length of database: 8,780,291
effective search space: 1036074338
effective search space used: 1036074338
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC139841.10


