

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139747.6 + phase: 0 /pseudo
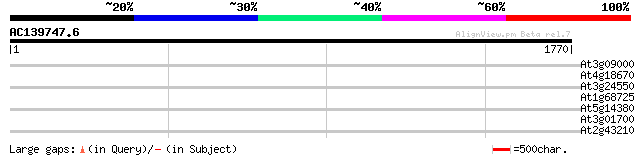
(1770 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g09000 unknown protein 35 0.30
At4g18670 extensin-like protein 35 0.51
At3g24550 protein kinase, putative 32 3.3
At1g68725 putative arabinogalactan protein AGP19 32 4.3
At5g14380 agp6 31 5.6
At3g01700 arabinogalactan-protein AGP11 31 5.6
At2g43210 unknown protein 30 9.6
>At3g09000 unknown protein
Length = 541
Score = 35.4 bits (80), Expect = 0.30
Identities = 34/107 (31%), Positives = 45/107 (41%), Gaps = 22/107 (20%)
Query: 3 PSTTILPPTTIPTKTTATTPSSGSSQSAESKATI*PLANDLWSIAPFFTCTESGPNNT-- 60
P+ PTT ++ T S+ S + S+AT+ + AP T T SG +
Sbjct: 162 PTRRSTTPTTSTSRPVTTRASNSRSSTPTSRATLTAARATTSTAAPRTTTTSSGSARSAT 221
Query: 61 -TSSHSRPS-------------PTLVPS---GPSLY---LPSRGTRP 87
T S+ RPS PT PS GPS+ PSRGT P
Sbjct: 222 PTRSNPRPSSASSKKPVSRPATPTRRPSTPTGPSIVSSKAPSRGTSP 268
>At4g18670 extensin-like protein
Length = 839
Score = 34.7 bits (78), Expect = 0.51
Identities = 26/79 (32%), Positives = 35/79 (43%), Gaps = 15/79 (18%)
Query: 9 PPTTIPTKTTATTPSSGSSQSAESKATI*PLANDLWSIAPFFTCTESGPNNTTSSHSRPS 68
PP +P+ T TPS G S + S + P+ S P + S PS
Sbjct: 406 PPVVVPSPPT--TPSPGGSPPSPSISPSPPIT------------VPSPPTTPSPGGSPPS 451
Query: 69 PTLVPSGPSLYLPSRGTRP 87
P++VPS PS PS G+ P
Sbjct: 452 PSIVPSPPST-TPSPGSPP 469
>At3g24550 protein kinase, putative
Length = 652
Score = 32.0 bits (71), Expect = 3.3
Identities = 28/86 (32%), Positives = 37/86 (42%), Gaps = 7/86 (8%)
Query: 4 STTILPPTTIPTKTTATTPSSGSSQSAESKATI*PLANDLWSIAPFFTCTESGPNNTTSS 63
STT PP + TTPSS S + +T P ++ L P S P + T
Sbjct: 22 STTTTPPPAASSPPPTTTPSS-PPPSPSTNSTSPPPSSPLPPSLP----PPSPPGSLTPP 76
Query: 64 HSRPSPT--LVPSGPSLYLPSRGTRP 87
+PSP+ + PS PS PS P
Sbjct: 77 LPQPSPSAPITPSPPSPTTPSNPRSP 102
>At1g68725 putative arabinogalactan protein AGP19
Length = 248
Score = 31.6 bits (70), Expect = 4.3
Identities = 26/89 (29%), Positives = 36/89 (40%), Gaps = 7/89 (7%)
Query: 3 PSTTILPPTTIPTKTTATTPSSGSSQSAESKAT----I*PLANDLWSIAPFFTCTESGPN 58
P TT PPTT T TTP ++Q S T + P + +AP + + P
Sbjct: 40 PPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPPAVTPTSPPAPKVAPVIS-PATPPP 98
Query: 59 NTTSSHSRPSPTLVPSGPSLYLPSRGTRP 87
S +PT+ P P + P T P
Sbjct: 99 QPPQSPPASAPTVSP--PPVSPPPAPTSP 125
>At5g14380 agp6
Length = 150
Score = 31.2 bits (69), Expect = 5.6
Identities = 28/79 (35%), Positives = 37/79 (46%), Gaps = 8/79 (10%)
Query: 3 PSTTILPPTTIPTKTTATTP--SSGSSQSAESKATI*PLANDLWSIAPFFTCTESGPNNT 60
P+ T P+ PTK A P SS SS A S A P+ D +S + + ++S T
Sbjct: 42 PTATTKAPSA-PTKAPAAAPKSSSASSPKASSPAAEGPVPEDDYSAS---SPSDSAEAPT 97
Query: 61 TSSHSRPSP--TLVPSGPS 77
SS P+P T GPS
Sbjct: 98 VSSPPAPTPDSTSAADGPS 116
>At3g01700 arabinogalactan-protein AGP11
Length = 136
Score = 31.2 bits (69), Expect = 5.6
Identities = 20/73 (27%), Positives = 30/73 (40%)
Query: 10 PTTIPTKTTATTPSSGSSQSAESKATI*PLANDLWSIAPFFTCTESGPNNTTSSHSRPSP 69
PT PTK+ P++ SA + P+A + + + S + S P+P
Sbjct: 28 PTASPTKSPTKAPAAAPKSSAAAPKASSPVAEEPTPEDDYSAASPSDSAEAPTVSSPPAP 87
Query: 70 TLVPSGPSLYLPS 82
T GPS PS
Sbjct: 88 TPEADGPSSDGPS 100
>At2g43210 unknown protein
Length = 531
Score = 30.4 bits (67), Expect = 9.6
Identities = 13/34 (38%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query: 4 STTILPPTTIPTKTTATTPSSGSS-QSAESKATI 36
S+ +LPP ++P +PS+ SS Q +E+K+T+
Sbjct: 146 SSVVLPPGSVPLDAAVASPSTASSVQPSETKSTV 179
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.359 0.158 0.619
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,591,437
Number of Sequences: 26719
Number of extensions: 1389261
Number of successful extensions: 5260
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 0
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 5235
Number of HSP's gapped (non-prelim): 21
length of query: 1770
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1657
effective length of database: 8,299,349
effective search space: 13752021293
effective search space used: 13752021293
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC139747.6


