

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139747.1 - phase: 0 /pseudo
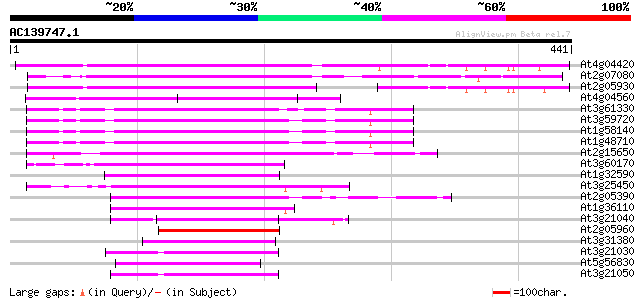
(441 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g04420 putative transposon protein 202 3e-52
At2g07080 putative gag-protease polyprotein 148 5e-36
At2g05930 copia-like retroelement pol polyprotein 145 4e-35
At4g04560 putative transposon protein 121 9e-28
At3g61330 copia-type polyprotein 111 7e-25
At3g59720 copia-type reverse transcriptase-like protein 110 2e-24
At1g58140 hypothetical protein 110 2e-24
At1g48710 hypothetical protein 110 2e-24
At2g15650 putative retroelement pol polyprotein 100 1e-21
At3g60170 putative protein 90 2e-18
At1g32590 hypothetical protein, 5' partial 86 3e-17
At3g25450 hypothetical protein 78 1e-14
At2g05390 putative retroelement pol polyprotein 77 1e-14
At1g36110 hypothetical protein 63 3e-10
At3g21040 hypothetical protein 62 6e-10
At2g05960 putative retroelement pol polyprotein 61 1e-09
At3g31380 hypothetical protein 59 5e-09
At3g21030 hypothetical protein 56 3e-08
At5g56830 unknown protein 55 8e-08
At3g21050 hypothetical protein 55 1e-07
>At4g04420 putative transposon protein
Length = 1008
Score = 202 bits (514), Expect = 3e-52
Identities = 138/476 (28%), Positives = 247/476 (50%), Gaps = 51/476 (10%)
Query: 5 KEGGFVNTPPLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLK 64
KE V +L+ NY +WK +M ++ + + IA GW+ PV DG +VLK
Sbjct: 5 KEFVAVGKTIMLEKGNYGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGE--DVLK 62
Query: 65 P*EEWTAAEDELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKS 124
++W AE+ A NS+AL+ +FN V++N FK I+ C AK+ + L A+EGT+ VK
Sbjct: 63 TKDQWNDAEEAKAKANSRALSLIFNFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKR 122
Query: 125 AKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFD 184
++ +L +++ENL M + E+I+++ I IA+ + G+K D+KLV+K+LR LP RF+
Sbjct: 123 SRIDMLASQFENLSMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFE 182
Query: 185 MKVTAIEEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDD 244
K TA+ + D S +E++G LQ +E+ + S KG+A A+S + +E+Q
Sbjct: 183 SKRTAMGTSLDTDSIDFEEVVGMLQAYELEITSGKGGYSKGLALAASAKKNEIQ------ 236
Query: 245 EDMTESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKM----------VRTDE 294
++ ++++++ + F + +++ +K+ Q D ++++ ++ +
Sbjct: 237 -ELKDTMSMMAKDFSRAMRRVEKKGFGRNQGTDRYRDRSSKRDEIQCHECQGYGHIKAEC 295
Query: 295 KNFQYKGVQCHECEGYGHIKIECASFLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALI 354
+ + K ++C EC G GH K +C K KS + S S+ D + GD E + IK + +
Sbjct: 296 PSLKRKDLKCSECNGLGHTKFDCVGSKSKPDKSCS-SESESDSNDGDSE-DYIKGFVSFV 353
Query: 355 GRV-------FSDAESCSEDLAYDE---------LVVSYKRLNDKNTDICKQ----LEEQ 394
G + S+A+ ED + DE + +++L D + K+ LEE+
Sbjct: 354 GIIEEKDESSDSEADGEDEDNSADEDSDIEKDVNINEEFRKLYDSWLMLSKEKVAWLEEK 413
Query: 395 ---KNITNNLEEERVGYLEKNSEL-------NSEVRMLNSQLSNVMKQVKMMAART 440
+ +T L+ E +KNSEL + R L+ +LS+ K + M+ + T
Sbjct: 414 LKVQELTEKLKGELTAANQKNSELIQKCSVAEEKNRELSQELSDTRKNIHMLNSGT 469
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 148 bits (374), Expect = 5e-36
Identities = 124/427 (29%), Positives = 200/427 (46%), Gaps = 53/427 (12%)
Query: 15 LLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAED 74
LLD Y YWK M+ ++RG E DG + KP WTA E
Sbjct: 15 LLDTKRYGYWKVCMT------------QIIRGQE-------DG--FKITKPKANWTAEEK 53
Query: 75 ELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKY 134
+ N++A+ A+FN VD++ FKLI+ C AK + L+ +HEGT+ VK + + T++
Sbjct: 54 LQSKFNARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSHEGTSSVKRTRLDHIATQF 113
Query: 135 ENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEAR 194
E L+M DE I + I +AN E G+ D+KLV+K+LR LP +F + A
Sbjct: 114 EYLKMEPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPKFAAHKAVMRVAG 173
Query: 195 DISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLL 254
+ +L+G L+ E+ + K K IAF + ++ Q ++ + + LL
Sbjct: 174 NTDKISFVDLVGMLKLEEMKADQDKVKPSKNIAFNADQGSEQFQ-------EIKDGMALL 226
Query: 255 GRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNF-QYKGVQCHECEGYGHI 313
R F K +K R ID + S+ + R++ + + K +QC+EC G+GHI
Sbjct: 227 ARNFGKALK-------------RVEIDGERSRGRFSRSENDDLRKKKEIQCYECGGFGHI 273
Query: 314 KIECASFLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSED------ 367
K EC + K K+ + +K + +S +LI FSD+ES E
Sbjct: 274 KPECP--ITKRKEMKCLKCKGVGHTKFECPNKSKLKEKSLIS--FSDSESDDEGEELLNF 329
Query: 368 LAYDELVVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLS 427
+A+ S K ++D ++D C + K+ L + V + +L E L ++L+
Sbjct: 330 VAFMASSDSSKFMSDTDSD-CDEELNPKDKYRVLYDSWVQLSKDKLKLVKEKLTLEAKLA 388
Query: 428 NVMKQVK 434
NV + K
Sbjct: 389 NVSTEDK 395
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 145 bits (366), Expect = 4e-35
Identities = 79/227 (34%), Positives = 135/227 (58%), Gaps = 2/227 (0%)
Query: 15 LLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAED 74
+L+ NY +WK +M ++ + + IA GW+ PV DG +VLK ++W AE+
Sbjct: 27 ILEKGNYGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGE--DVLKTEDQWNDAEE 84
Query: 75 ELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKY 134
A NS+AL+ +FN V++N FK I+ C AK+ + L A+EGT+ VK ++ +L +++
Sbjct: 85 AKATANSRALSLIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQF 144
Query: 135 ENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEAR 194
ENL M + E+I+++ I IA+ + G+K D+KLV+K+LR LP RF+ K TA+ +
Sbjct: 145 ENLTMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSL 204
Query: 195 DISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQ 241
D +S +E++G Q +E+ + S G A S++ +L+ ++
Sbjct: 205 DTNSIDFEEVVGMFQAYELEITSGKGGYGHIKAECPSLKRKDLKCSE 251
Score = 54.3 bits (129), Expect = 1e-07
Identities = 48/181 (26%), Positives = 87/181 (47%), Gaps = 32/181 (17%)
Query: 290 VRTDEKNFQYKGVQCHECEGYGHIKIECASFLKK*KKSLTVSWSDDDGSKGDGERESIKH 349
++ + + + K ++C EC+G GHIK +C K +S + S S+ D + GD E + IK
Sbjct: 235 IKAECPSLKRKDLKCSECKGLGHIKFDCVGSKSKPDRSCS-SESESDSNDGDSE-DYIKG 292
Query: 350 VAALIGRV-------FSDAESCSEDLAYDE---------LVVSYKRLNDKNTDICKQ--- 390
+ +G + S+A+ ED + DE + +++L D + K+
Sbjct: 293 FVSFVGIIEEKDESSDSEADGEDEDNSADEDSDIEKDVKINEEFRKLYDSWLMLSKEKVA 352
Query: 391 -LEEQ---KNITNNLEEERVGYLEKNSELNSEV-------RMLNSQLSNVMKQVKMMAAR 439
LEE+ + +T L+ E +KNSEL + R L+ +LS+ K++ M+ +
Sbjct: 353 WLEEKLKVQELTEKLKGELTAANQKNSELTQKCSVAEEKNRELSQELSDTRKKIHMLNSG 412
Query: 440 T 440
T
Sbjct: 413 T 413
>At4g04560 putative transposon protein
Length = 590
Score = 121 bits (303), Expect = 9e-28
Identities = 73/248 (29%), Positives = 125/248 (49%), Gaps = 5/248 (2%)
Query: 13 PPLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAA 72
P LD +Y YWK + ++ +D AV GW P T D V K EW A
Sbjct: 345 PLKLDAEHYGYWKVLIKRSIQSIDMDAWFAVEDGWMPPTT--KDAKRDIVSKSRTEWIAD 402
Query: 73 EDELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTT 132
E A NS+AL+ +F + +N F ++ C+ AK+ EIL+++ E T VK + +L +
Sbjct: 403 EKTAANHNSQALSVIFGSLLRNKFTQVQGCLSAKEVWEILQVSFECTNNVKRTRLDMLAS 462
Query: 133 KYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEE 192
++ENL M +ES+ D++ + I G+ D+K+V+K LRSLP +F +AI+
Sbjct: 463 EFENLTMEAEESVDDFNGKLSSITQEAVVLGKTYKDKKMVKKFLRSLPDKFQSHKSAIDV 522
Query: 193 ARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLT 252
+ + K D+++G +Q ++ K + + ++E D+ V ++ +SL
Sbjct: 523 SLNSDQLKFDQVVGMMQAYD---TDKEEILNSYATYFGAIEDDDHTVEEDAQMGTIKSLI 579
Query: 253 LLGRQFKK 260
L+ +K
Sbjct: 580 LIQSDSEK 587
Score = 58.2 bits (139), Expect = 9e-09
Identities = 29/94 (30%), Positives = 54/94 (56%)
Query: 133 KYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEE 192
+YENL+M + ++I + ++++ N GE+ SD ++V+KIL SLPKRFD+ V +++
Sbjct: 97 EYENLKMKESDNINTFMTKLIEMGNQLRVHGEEKSDYQIVQKILISLPKRFDIIVAMMKQ 156
Query: 193 ARDISSFKVDELIGSLQNFEITVNSKNDKKGKGI 226
+D++S + + + KK KG+
Sbjct: 157 TKDLTSLSAGKWCDVCERKNHNESDCWMKKNKGV 190
>At3g61330 copia-type polyprotein
Length = 1352
Score = 111 bits (278), Expect = 7e-25
Identities = 81/310 (26%), Positives = 151/310 (48%), Gaps = 32/310 (10%)
Query: 14 PLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAE 73
P+L NYD W RM L D V +G+ P +G+++ K +
Sbjct: 11 PVLTKSNYDNWSLRMKAILGAHDVWE--IVEKGFIEPEN---EGSLSQTQKDGLRDSRKR 65
Query: 74 DELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTK 133
D+ KAL ++ +D++ F+ + + AK+ E L+ +++G +VK + Q L +
Sbjct: 66 DK------KALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 134 YENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEA 193
+E L+M + E + DY +L + N+ + GEK+ D +++ K+LRSL +F+ VT IEE
Sbjct: 120 FEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEET 179
Query: 194 RDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTL 253
+D+ + +++L+GSLQ +E + KK + IA ++ +Q+ +E++ +
Sbjct: 180 KDLEAMTIEQLLGSLQAYE-----EKKKKKEDIA----EQVLNMQITKEENGQSYQ---- 226
Query: 254 LGRQFKKIVKQYDKRPRSIGQNIRPNIDN------QPSKEKMVRTDEKNFQYKGVQCHEC 307
R+ V+ + G+ RP+ DN S+ + + + V+C+ C
Sbjct: 227 --RRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNC 284
Query: 308 EGYGHIKIEC 317
+GH EC
Sbjct: 285 GKFGHYASEC 294
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 110 bits (274), Expect = 2e-24
Identities = 79/310 (25%), Positives = 147/310 (46%), Gaps = 32/310 (10%)
Query: 14 PLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAE 73
P+L NYD W RM L D V +G+ P +G+++ K +
Sbjct: 11 PVLTKSNYDNWSLRMKAILGAHDVWE--IVEKGFIEPEN---EGSLSQTQKDGLRDSRKR 65
Query: 74 DELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTK 133
D+ KAL ++ +D++ F+ + + AK+ E L+ +++G +VK + Q L +
Sbjct: 66 DK------KALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 134 YENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEA 193
+E L+M + E + DY +L + N+ + GEK+ D +++ K+LRSL +F+ VT IEE
Sbjct: 120 FEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEET 179
Query: 194 RDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTL 253
+D+ + +++L+GSLQ +E K D ++ +Q+ +E++ +
Sbjct: 180 KDLEAMTIEQLLGSLQAYEEKKKKKED---------IVEQVLNMQITKEENGQSYQ---- 226
Query: 254 LGRQFKKIVKQYDKRPRSIGQNIRPNIDN------QPSKEKMVRTDEKNFQYKGVQCHEC 307
R+ V+ + G+ RP+ DN S+ + + + V+C+ C
Sbjct: 227 --RRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNC 284
Query: 308 EGYGHIKIEC 317
+GH EC
Sbjct: 285 GKFGHYASEC 294
>At1g58140 hypothetical protein
Length = 1320
Score = 110 bits (274), Expect = 2e-24
Identities = 79/310 (25%), Positives = 147/310 (46%), Gaps = 32/310 (10%)
Query: 14 PLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAE 73
P+L NYD W RM L D V +G+ P +G+++ K +
Sbjct: 11 PVLTKSNYDNWSLRMKAILGAHDVWE--IVEKGFIEPEN---EGSLSQTQKDGLRDSRKR 65
Query: 74 DELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTK 133
D+ KAL ++ +D++ F+ + + AK+ E L+ +++G +VK + Q L +
Sbjct: 66 DK------KALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 134 YENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEA 193
+E L+M + E + DY +L + N+ + GEK+ D +++ K+LRSL +F+ VT IEE
Sbjct: 120 FEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEET 179
Query: 194 RDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTL 253
+D+ + +++L+GSLQ +E K D ++ +Q+ +E++ +
Sbjct: 180 KDLEAMTIEQLLGSLQAYEEKKKKKED---------IVEQVLNMQITKEENGQSYQ---- 226
Query: 254 LGRQFKKIVKQYDKRPRSIGQNIRPNIDN------QPSKEKMVRTDEKNFQYKGVQCHEC 307
R+ V+ + G+ RP+ DN S+ + + + V+C+ C
Sbjct: 227 --RRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNC 284
Query: 308 EGYGHIKIEC 317
+GH EC
Sbjct: 285 GKFGHYASEC 294
>At1g48710 hypothetical protein
Length = 1352
Score = 110 bits (274), Expect = 2e-24
Identities = 79/310 (25%), Positives = 147/310 (46%), Gaps = 32/310 (10%)
Query: 14 PLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAE 73
P+L NYD W RM L D V +G+ P +G+++ K +
Sbjct: 11 PVLTKSNYDNWSLRMKAILGAHDVWE--IVEKGFIEPEN---EGSLSQTQKDGLRDSRKR 65
Query: 74 DELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTK 133
D+ KAL ++ +D++ F+ + + AK+ E L+ +++G +VK + Q L +
Sbjct: 66 DK------KALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 134 YENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEA 193
+E L+M + E + DY +L + N+ + GEK+ D +++ K+LRSL +F+ VT IEE
Sbjct: 120 FEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEET 179
Query: 194 RDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTL 253
+D+ + +++L+GSLQ +E K D ++ +Q+ +E++ +
Sbjct: 180 KDLEAMTIEQLLGSLQAYEEKKKKKED---------IIEQVLNMQITKEENGQSYQ---- 226
Query: 254 LGRQFKKIVKQYDKRPRSIGQNIRPNIDN------QPSKEKMVRTDEKNFQYKGVQCHEC 307
R+ V+ + G+ RP+ DN S+ + + + V+C+ C
Sbjct: 227 --RRGGGQVRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNC 284
Query: 308 EGYGHIKIEC 317
+GH EC
Sbjct: 285 GKFGHYASEC 294
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 100 bits (250), Expect = 1e-21
Identities = 77/329 (23%), Positives = 143/329 (43%), Gaps = 41/329 (12%)
Query: 14 PLLDGLNYDYWKSRMSVFLK------FVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*E 67
P+ DG YD+W +M+ + V+ + ++ E P T A
Sbjct: 10 PIFDGEKYDFWSIKMATIFRTRKLWSVVEEGVPVEPVQAEETPETARAK----------- 58
Query: 68 EWTAAEDELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKF 127
+E ++ AL L V +F I +K+ ++LK ++G+ +V+ K
Sbjct: 59 ---TLREEAVTNDTMALQILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKL 115
Query: 128 QLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKV 187
Q L +YENL+M D+++I+ + ++ + GEK ++ +L++KIL SLP +FD V
Sbjct: 116 QSLRREYENLKMYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIV 175
Query: 188 TAIEEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDM 247
+ +E+ RD+ + + EL+G L+ E V ++ + +G + S + + +
Sbjct: 176 SVLEQTRDLDALTMSELLGILKAQEARVTAREESTKEGAFYVRSKGRESGFKQDNTNNRV 235
Query: 248 TESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHEC 307
+ G F K K ++ R EK D + ++C++C
Sbjct: 236 NQDKKWCG--FHKSSKHTEEECR----------------EKPKNDDHGKNKRSNIKCYKC 277
Query: 308 EGYGHIKIECASFLKK*KKSLTVSWSDDD 336
GH EC S K K+ V+ ++D
Sbjct: 278 GKIGHYANECRS---KNKERAHVTLEEED 303
>At3g60170 putative protein
Length = 1339
Score = 90.1 bits (222), Expect = 2e-18
Identities = 66/203 (32%), Positives = 101/203 (49%), Gaps = 11/203 (5%)
Query: 14 PLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAE 73
P DG YD+W M FL+ + + R E + G T V E +A
Sbjct: 13 PRFDGY-YDFWSMTMENFLRSRE------LWRLVEEGIPAIVVGT-TPVS---EAQRSAV 61
Query: 74 DELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTK 133
+E L + K N LF +D+ + + I +K E +K ++G+TKVK A+ Q L +
Sbjct: 62 EEAKLKDLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKE 121
Query: 134 YENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEA 193
+E L M + E I + L + N ++ GE + +V KILRSL +F+ V +IEE+
Sbjct: 122 FELLAMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEES 181
Query: 194 RDISSFKVDELIGSLQNFEITVN 216
D+S+ +DEL GSL E +N
Sbjct: 182 NDLSTLSIDELHGSLLVHEQRLN 204
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 86.3 bits (212), Expect = 3e-17
Identities = 51/138 (36%), Positives = 75/138 (53%)
Query: 75 ELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKY 134
E + + K N LF +DK + K I Q +KD E +K ++G +V+SA+ Q L +
Sbjct: 20 EKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSF 79
Query: 135 ENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEAR 194
E L M E+I Y +++I N + GE + D K+V KILR+L ++F V AIEE+
Sbjct: 80 EVLEMKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEESN 139
Query: 195 DISSFKVDELIGSLQNFE 212
+I VD L SL E
Sbjct: 140 NIKELTVDGLQSSLMVHE 157
>At3g25450 hypothetical protein
Length = 1343
Score = 77.8 bits (190), Expect = 1e-14
Identities = 62/260 (23%), Positives = 122/260 (46%), Gaps = 35/260 (13%)
Query: 14 PLLDGLNYDYWKSRMSVFLKFVDNKT*IAVLRGWEHPVTLYADGNMTNVLKP*EEWTAAE 73
P+L+ +NY W RM L+ V + W ++P +A+
Sbjct: 23 PMLNSVNYTVWTMRMEAVLR---------VHKLW-------------GTIEP----GSAD 56
Query: 74 DELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTK 133
+E N A LF + +++ + + + E +K + G +VK A+ Q L +
Sbjct: 57 EEK---NDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMAE 113
Query: 134 YENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLP-KRFDMKVTAIEE 192
++ L+M D E+I DY I +I + GE I + K+V+K L+SLP K++ V A+E+
Sbjct: 114 FDKLKMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALEQ 173
Query: 193 ARDISSFKVDELIGSLQNFEITV---NSKNDKKGKGIAFASSVELDELQVNQED--DEDM 247
D+ + +++ G ++ +E V + ++ +GK + +D+L+ +E+ ++++
Sbjct: 174 VLDLKTTTFEDIAGRIKTYEDRVWDDDDSHEDQGKLMTEVEEEVVDDLEEEEEEVINKEI 233
Query: 248 TESLTLLGRQFKKIVKQYDK 267
++ R K I Q K
Sbjct: 234 KAKSHVIDRLLKLIRLQEQK 253
Score = 29.6 bits (65), Expect = 3.5
Identities = 22/94 (23%), Positives = 43/94 (45%), Gaps = 7/94 (7%)
Query: 344 RESIKHVAALIGRVFSDAESCSEDLAYDELVVSYKRLNDKNTDICKQLEEQKNITNNLEE 403
R+ H+ A + +V + ED+A K D+ D E+Q + +EE
Sbjct: 161 RKKYIHIVAALEQVLDLKTTTFEDIAG-----RIKTYEDRVWDDDDSHEDQGKLMTEVEE 215
Query: 404 ERVGYLEKNSE--LNSEVRMLNSQLSNVMKQVKM 435
E V LE+ E +N E++ + + ++K +++
Sbjct: 216 EVVDDLEEEEEEVINKEIKAKSHVIDRLLKLIRL 249
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 77.4 bits (189), Expect = 1e-14
Identities = 72/270 (26%), Positives = 122/270 (44%), Gaps = 41/270 (15%)
Query: 80 NSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRM 139
N A LF V ++ + + +K E +K + G +VK AK Q L +++ L M
Sbjct: 24 NDMARALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAKLQTLMAEFDRLNM 83
Query: 140 LDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLP-KRFDMKVTAIEEARDISS 198
D+E+I ++ I +I+ ES GE+I + K+V+K L+SLP K++ + A+E+ D+++
Sbjct: 84 KDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHIIAALEQILDLNT 143
Query: 199 FKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQ-VNQEDDEDMTESLTLLGRQ 257
++++G ++ +E V ++D S E +L N E D T GR
Sbjct: 144 TGFEDIVGRMKTYEDRVCDEDD---------SPEEQGKLMYANSESSYD-----TRGGRG 189
Query: 258 FKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIEC 317
R RS G+ Q K K++ C+ C+ GH EC
Sbjct: 190 --------RGRGRSSGRGRGGYGYQQRDKSKVI-------------CYRCDKTGHYASEC 228
Query: 318 ASFLKK*KKSLTVSWSDDDGSKGDGERESI 347
L K K+ +++D D E ES+
Sbjct: 229 LDRLLKLIKAQEQQQNNED----DDEIESL 254
>At1g36110 hypothetical protein
Length = 745
Score = 63.2 bits (152), Expect = 3e-10
Identities = 34/149 (22%), Positives = 78/149 (51%), Gaps = 4/149 (2%)
Query: 80 NSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRM 139
N+ A +F + +++ + AK + +K + G +VK A+ Q L ++E ++M
Sbjct: 57 NTMARGLIFQSIPESLTLQVGTLATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMKM 116
Query: 140 LDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLP-KRFDMKVTAIEEARDISS 198
+ E I + + ++A ++ G I KLV+K L +LP +++ + ++E+ D+++
Sbjct: 117 KESEKIDVFAGRLAELATRSDALGSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLNN 176
Query: 199 FKVDELIGSLQNFEITV---NSKNDKKGK 224
++++G ++ +E V + D +GK
Sbjct: 177 TSFEDIVGRIKVYEERVWDGEEQEDDQGK 205
>At3g21040 hypothetical protein
Length = 534
Score = 62.0 bits (149), Expect = 6e-10
Identities = 46/189 (24%), Positives = 88/189 (46%), Gaps = 6/189 (3%)
Query: 80 NSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRM 139
++KAL + + + ++F+ +K+ ++LK +G ++ K + L + ENL M
Sbjct: 68 DNKALKIMQSALPDSVFRKTISVASSKELWDLLK---KGNDNEEAKKLRRLEKQLENLTM 124
Query: 140 LDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDISSF 199
+ E ++ Y + I F G ISDEK++ K+L SLP+ +D +T ++E +
Sbjct: 125 YEGERMKSYLKRVEKIIEEFFVWGNPISDEKVIAKLLTSLPRPYDDSITVLKEFMTLPDL 184
Query: 200 KVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTL--LGRQ 257
+L+ + + F + + K I D L D + + L L +Q
Sbjct: 185 THRDLLKAFEMFGSNPKTMPQELMKFINILRKAHSDRLPCRGYDKNNPIKDAKLLRLEKQ 244
Query: 258 FKKIVKQYD 266
F+K++ YD
Sbjct: 245 FEKLM-MYD 252
Score = 58.2 bits (139), Expect = 9e-09
Identities = 29/96 (30%), Positives = 50/96 (51%)
Query: 116 HEGTTKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKI 175
++ +K AK L ++E L M D ES+ Y +LD F ++G +SD+ ++ K+
Sbjct: 227 YDKNNPIKDAKLLRLEKQFEKLMMYDGESMDSYLERVLDTIEQFRASGNPLSDDDVIAKL 286
Query: 176 LRSLPKRFDMKVTAIEEARDISSFKVDELIGSLQNF 211
L SL +D + +EE ++ +LIG L+ F
Sbjct: 287 LTSLSWPYDDAIPVLEELMNLPDMTPHDLIGVLEMF 322
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 60.8 bits (146), Expect = 1e-09
Identities = 30/96 (31%), Positives = 58/96 (60%), Gaps = 1/96 (1%)
Query: 118 GTTKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILR 177
G +VK AK + L +++ L+M D+E+I + + +I+ S GE I + K+V+K L+
Sbjct: 56 GAERVKEAKLKTLMAEFDKLKMKDNETIDECAGRLSEISTKSTSLGEDIEETKVVKKFLK 115
Query: 178 SLP-KRFDMKVTAIEEARDISSFKVDELIGSLQNFE 212
SLP K++ V A+E+ D+ + +++G ++ +E
Sbjct: 116 SLPTKKYIHIVAALEQVLDLKNTTFKDIVGRIKTYE 151
>At3g31380 hypothetical protein
Length = 262
Score = 58.9 bits (141), Expect = 5e-09
Identities = 29/106 (27%), Positives = 59/106 (55%), Gaps = 1/106 (0%)
Query: 105 AKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGE 164
AK+ + +K H G +VK A+ Q L +E ++M + E I D+ + +++ G
Sbjct: 24 AKEVWDSIKTRHVGAERVKEARVQTLMADFEKMKMKEAEKIDDFAGRLSELSTKSAPLGV 83
Query: 165 KISDEKLVRKILRSLP-KRFDMKVTAIEEARDISSFKVDELIGSLQ 209
I KLV+K L SLP K++ + ++E+ D+++ ++++G ++
Sbjct: 84 NIEVPKLVKKFLNSLPRKKYIHIIASLEQVLDLNNTTFEDIVGCMK 129
>At3g21030 hypothetical protein
Length = 411
Score = 56.2 bits (134), Expect = 3e-08
Identities = 35/136 (25%), Positives = 69/136 (50%), Gaps = 4/136 (2%)
Query: 76 LALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYE 135
L + ++KAL + + + ++F+ AK+ ++LK ++ K AK + L ++E
Sbjct: 64 LVVKDNKALKIMQSSLPDSVFRKTISIASAKELWDLLKKGND----TKEAKLRRLEKQFE 119
Query: 136 NLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARD 195
NL M + E ++ Y + I G ISD+K++ K+L SLP+ +D V ++E
Sbjct: 120 NLMMYEGERMKLYLKRLEKIIEELFVLGNPISDDKVIAKLLTSLPRSYDDSVPVLKEFMT 179
Query: 196 ISSFKVDELIGSLQNF 211
+ +L+ + + F
Sbjct: 180 LPELTHRDLLKAFEMF 195
>At5g56830 unknown protein
Length = 242
Score = 55.1 bits (131), Expect = 8e-08
Identities = 31/114 (27%), Positives = 58/114 (50%)
Query: 84 LNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRMLDDE 143
L+ + + + +F I +K+ EIL + G ++ K L +Y N M D E
Sbjct: 14 LSLIQSGLSTEIFSWIAYATSSKEAWEILMVRSVGGPIFQARKLLDLKKEYNNTTMSDAE 73
Query: 144 SIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDIS 197
++ ++ +L++ +S G K+ ++ LV+KIL SLP RFD + + E ++
Sbjct: 74 TVHEFTGKLLELVFRMKSCGWKVEEKILVKKILSSLPPRFDKVLVEVAEKLSVN 127
>At3g21050 hypothetical protein
Length = 420
Score = 54.7 bits (130), Expect = 1e-07
Identities = 33/132 (25%), Positives = 67/132 (50%), Gaps = 4/132 (3%)
Query: 80 NSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRM 139
++KAL + + + ++F+ AK+ ++LK ++ K AK + L ++E L M
Sbjct: 68 DNKALKIMQSSLPDSVFRKTISIASAKELWDLLKKGND----TKEAKLRRLEKQFEKLMM 123
Query: 140 LDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDISSF 199
+ E + Y + +I FE G ISD+K++ K+L SL +D + ++E +
Sbjct: 124 YEGEPMDLYLKRVEEITERFEVLGNPISDDKVITKLLTSLSWPYDDSIPVLKEFMTLPDL 183
Query: 200 KVDELIGSLQNF 211
+ +L+ + + F
Sbjct: 184 TLRDLLKAFELF 195
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,581,671
Number of Sequences: 26719
Number of extensions: 417693
Number of successful extensions: 1827
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 1691
Number of HSP's gapped (non-prelim): 158
length of query: 441
length of database: 11,318,596
effective HSP length: 102
effective length of query: 339
effective length of database: 8,593,258
effective search space: 2913114462
effective search space used: 2913114462
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139747.1


