

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139745.7 + phase: 0
(240 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
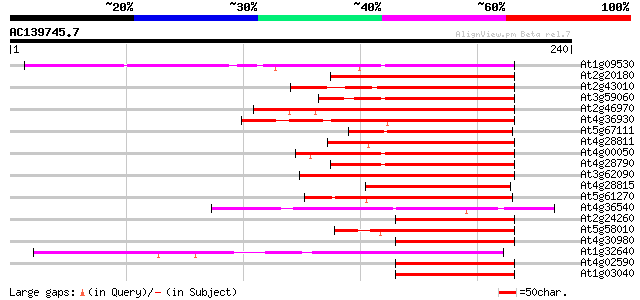
Sequences producing significant alignments: (bits) Value
At1g09530 putative transcription factor BHLH8 111 3e-25
At2g20180 putative bHLH transcription factor (bHLH015) 93 1e-19
At2g43010 putative transcription factor BHLH9 93 1e-19
At3g59060 putative bHLH transcription factor (bHLH065) 91 7e-19
At2g46970 PIF3 like basic Helix Loop Helix protein (PIL1) 89 2e-18
At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATU... 86 1e-17
At5g67111 putative bHLH transcription factor (bHLH073/ALCATRAZ) 84 5e-17
At4g28811 AtbHLH119 80 7e-16
At4g00050 bHLH like transcription factor (bHLH016) 77 6e-15
At4g28790 bHLH transcription factor like protein (bHLH023) 76 1e-14
At3g62090 PIF3 like basic Helix Loop Helix protein 2 (PIL2) 75 3e-14
At4g28815 AtbHLH127 75 4e-14
At5g61270 bHLH transcription factor like protein 70 1e-12
At4g36540 putative bHLH transcription factor (bHLH058) 67 6e-12
At2g24260 putative bHLH transcription factor (bHLH066) 67 6e-12
At5g58010 putative bHLH transcription factor (bHLH082) 66 1e-11
At4g30980 putative bHLH transcription factor (bHLH069) 66 1e-11
At1g32640 transcription factor like protein, BHLH6 66 1e-11
At4g02590 putative bHLH transcription factor (bHLH059) 65 3e-11
At1g03040 putative transcription factor (BHLH7) 64 7e-11
>At1g09530 putative transcription factor BHLH8
Length = 524
Score = 111 bits (278), Expect = 3e-25
Identities = 79/212 (37%), Positives = 116/212 (54%), Gaps = 9/212 (4%)
Query: 7 KNGKVNFSNFSIPAVFLKSNQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSGYERSRE 66
K VNFS+F PA F K+ + + + K+ Q + L A + S
Sbjct: 194 KPSLVNFSHFLRPATFAKTTNNNLHDTKEKSPQSPPNVFQTRV-LGAKDSEDKVLNESVA 252
Query: 67 LLPTVDEHSEAAASHTNAPRIRGKRKASAINLCNAQKPSSVCSLEA-SNDLNFGVRKSHE 125
D S + + + KA +C++ + SL+ S + +++ H
Sbjct: 253 SATPKDNQKACLISEDSCRKDQESEKAV---VCSSVGSGN--SLDGPSESPSLSLKRKHS 307
Query: 126 DTDDSPYLSDNDEETQENIVKEK-PVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRAL 184
+ D S++ EE + KE P R G KRS R+A+VHNLSER+RRD+INEK+RAL
Sbjct: 308 NIQDIDCHSEDVEEESGDGRKEAGPSRTGLGSKRS-RSAEVHNLSERRRRDRINEKMRAL 366
Query: 185 KELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ELIPNCNK+DKASMLD+AI+YLK+L+LQ+Q+
Sbjct: 367 QELIPNCNKVDKASMLDEAIEYLKSLQLQVQI 398
>At2g20180 putative bHLH transcription factor (bHLH015)
Length = 478
Score = 93.2 bits (230), Expect = 1e-19
Identities = 44/79 (55%), Positives = 61/79 (76%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
+ET+ + K R + R A+VHNLSERKRRD+INE+++AL+ELIP CNK DKA
Sbjct: 261 DETESRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCNKSDKA 320
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+Y+K+L+LQ+Q+
Sbjct: 321 SMLDEAIEYMKSLQLQIQM 339
>At2g43010 putative transcription factor BHLH9
Length = 430
Score = 92.8 bits (229), Expect = 1e-19
Identities = 53/96 (55%), Positives = 69/96 (71%), Gaps = 9/96 (9%)
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
RK TD+S LSD I + R G+ R R A+VHNLSER+RRD+INE+
Sbjct: 226 RKRINHTDESVSLSDA-------IGNKSNQRSGSN--RRSRAAEVHNLSERRRRDRINER 276
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
++AL+ELIP+C+K DKAS+LD+AIDYLK+L+LQLQV
Sbjct: 277 MKALQELIPHCSKTDKASILDEAIDYLKSLQLQLQV 312
>At3g59060 putative bHLH transcription factor (bHLH065)
Length = 442
Score = 90.5 bits (223), Expect = 7e-19
Identities = 46/84 (54%), Positives = 66/84 (77%), Gaps = 5/84 (5%)
Query: 133 LSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCN 192
L+ D++T N K + +RS R A+VHNLSER+RRD+INE+++AL+ELIP+C+
Sbjct: 233 LTSTDDQTMGN----KSSQRSGSTRRS-RAAEVHNLSERRRRDRINERMKALQELIPHCS 287
Query: 193 KMDKASMLDDAIDYLKTLKLQLQV 216
+ DKAS+LD+AIDYLK+L++QLQV
Sbjct: 288 RTDKASILDEAIDYLKSLQMQLQV 311
>At2g46970 PIF3 like basic Helix Loop Helix protein (PIL1)
Length = 416
Score = 89.4 bits (220), Expect = 2e-18
Identities = 48/115 (41%), Positives = 77/115 (66%), Gaps = 3/115 (2%)
Query: 105 SSVCSLEASNDLNF-GVRKSHEDTDD--SPYLSDNDEETQENIVKEKPVREGNRVKRSYR 161
SS S S DL+ +++ + D ++ S YLS+N ++ ++ + R V + R
Sbjct: 170 SSKFSRGTSRDLSCCSLKRKYGDIEEEESTYLSNNSDDESDDAKTQVHARTRKPVTKRKR 229
Query: 162 NAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ +VH L ERKRRD+ N+K+RAL++L+PNC K DKAS+LD+AI Y++TL+LQ+Q+
Sbjct: 230 STEVHKLYERKRRDEFNKKMRALQDLLPNCYKDDKASLLDEAIKYMRTLQLQVQM 284
>At4g36930 putative bHLH transcription factor (AtbHLH024) / SPATULA
(SPT)
Length = 373
Score = 86.3 bits (212), Expect = 1e-17
Identities = 56/121 (46%), Positives = 75/121 (61%), Gaps = 12/121 (9%)
Query: 100 NAQKPSSVCSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRS 159
N Q SS + +S+ V S +TD+ S EE E +V E P + RS
Sbjct: 140 NVQGNSSGTRVSSSS-----VGASGNETDEYDCES---EEGGEAVVDEAPSSKSGPSSRS 191
Query: 160 Y----RNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQ 215
R A+VHNLSE++RR +INEK++AL+ LIPN NK DKASMLD+AI+YLK L+LQ+Q
Sbjct: 192 SSKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPNSNKTDKASMLDEAIEYLKQLQLQVQ 251
Query: 216 V 216
+
Sbjct: 252 M 252
>At5g67111 putative bHLH transcription factor (bHLH073/ALCATRAZ)
Length = 210
Score = 84.3 bits (207), Expect = 5e-17
Identities = 42/70 (60%), Positives = 57/70 (81%), Gaps = 1/70 (1%)
Query: 146 KEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAID 205
K ++ N +KR+ +A+ HNLSE+KRR KINEK++AL++LIPN NK DKASMLD+AI+
Sbjct: 79 KRSGAKQRNSLKRNI-DAQFHNLSEKKRRSKINEKMKALQKLIPNSNKTDKASMLDEAIE 137
Query: 206 YLKTLKLQLQ 215
YLK L+LQ+Q
Sbjct: 138 YLKQLQLQVQ 147
>At4g28811 AtbHLH119
Length = 470
Score = 80.5 bits (197), Expect = 7e-16
Identities = 37/82 (45%), Positives = 61/82 (74%), Gaps = 2/82 (2%)
Query: 137 DEETQENIVKEKPVRE--GNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKM 194
D++ +E I + + E G+ ++ R A +HNLSER+RR++INE+++ L+EL+P C K
Sbjct: 254 DKKREETIAEIQGTEEAHGSTSRKRSRAADMHNLSERRRRERINERMKTLQELLPRCRKT 313
Query: 195 DKASMLDDAIDYLKTLKLQLQV 216
DK SML+D I+Y+K+L+LQ+Q+
Sbjct: 314 DKVSMLEDVIEYVKSLQLQIQM 335
>At4g00050 bHLH like transcription factor (bHLH016)
Length = 399
Score = 77.4 bits (189), Expect = 6e-15
Identities = 41/95 (43%), Positives = 64/95 (67%), Gaps = 2/95 (2%)
Query: 123 SHEDT-DDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKI 181
SH++T DD + + + ++ K+ + KRS R A +HN SERKRRDKIN+++
Sbjct: 175 SHDNTIDDHDSVCHSRPQMEDEEEKKAGGKSSVSTKRS-RAAAIHNQSERKRRDKINQRM 233
Query: 182 RALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ L++L+PN +K DKASMLD+ I+YLK L+ Q+ +
Sbjct: 234 KTLQKLVPNSSKTDKASMLDEVIEYLKQLQAQVSM 268
>At4g28790 bHLH transcription factor like protein (bHLH023)
Length = 413
Score = 76.3 bits (186), Expect = 1e-14
Identities = 39/79 (49%), Positives = 55/79 (69%), Gaps = 1/79 (1%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
EET + R+ KRS R A +H LSER+RR KINE ++AL+EL+P C K D++
Sbjct: 255 EETNVENQGTEEARDSTSSKRS-RAAIMHKLSERRRRQKINEMMKALQELLPRCTKTDRS 313
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLDD I+Y+K+L+ Q+Q+
Sbjct: 314 SMLDDVIEYVKSLQSQIQM 332
>At3g62090 PIF3 like basic Helix Loop Helix protein 2 (PIL2)
Length = 363
Score = 75.1 bits (183), Expect = 3e-14
Identities = 36/92 (39%), Positives = 61/92 (66%)
Query: 125 EDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRAL 184
ED++ S YLS + ++ ++ + P R + + RNA+ +N ER +R+ IN+K+R L
Sbjct: 152 EDSEGSMYLSSSLDDESDDARPQVPARTRKALVKRKRNAEAYNSPERNQRNDINKKMRTL 211
Query: 185 KELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ L+PN +K D SMLD+AI+Y+ L+LQ+Q+
Sbjct: 212 QNLLPNSHKDDNESMLDEAINYMTNLQLQVQM 243
>At4g28815 AtbHLH127
Length = 307
Score = 74.7 bits (182), Expect = 4e-14
Identities = 31/62 (50%), Positives = 51/62 (82%)
Query: 153 GNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKL 212
G+ ++ R A++HNL+ER+RR+KINE+++ L++LIP CNK K SML+D I+Y+K+L++
Sbjct: 142 GSTSRKRSRAAEMHNLAERRRREKINERMKTLQQLIPRCNKSTKVSMLEDVIEYVKSLEM 201
Query: 213 QL 214
Q+
Sbjct: 202 QI 203
>At5g61270 bHLH transcription factor like protein
Length = 366
Score = 70.1 bits (170), Expect = 1e-12
Identities = 39/91 (42%), Positives = 56/91 (60%), Gaps = 3/91 (3%)
Query: 127 TDDSPYLSDNDEETQENIVKEKPVR--EGNRVKRSYRNAKVHNLSERKRRDKINEKIRAL 184
T D Y E TQ+ E+ R G R R A +HN SER+RRD+IN+++R L
Sbjct: 131 TGDRDYFRSGSE-TQDTEGDEQETRGEAGRSNGRRGRAAAIHNESERRRRDRINQRMRTL 189
Query: 185 KELIPNCNKMDKASMLDDAIDYLKTLKLQLQ 215
++L+P +K DK S+LDD I++LK L+ Q+Q
Sbjct: 190 QKLLPTASKADKVSILDDVIEHLKQLQAQVQ 220
>At4g36540 putative bHLH transcription factor (bHLH058)
Length = 304
Score = 67.4 bits (163), Expect = 6e-12
Identities = 45/148 (30%), Positives = 80/148 (53%), Gaps = 9/148 (6%)
Query: 87 IRGKRKASAINLCNAQKPSSVCSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVK 146
++ + AI L N +KP + K+ ++T+ S N T+ +
Sbjct: 79 VKNNGHSRAITLQNKRKPEGKTEKREKKKI-----KAEDETEPSMKGKSNMSNTETSSEI 133
Query: 147 EKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKM-DKASMLDDAID 205
+KP R +R + H+L+ER RR+KI++K++ L++++P CNK+ KA MLD+ I+
Sbjct: 134 QKPDYIHVRARRGEATDR-HSLAERARREKISKKMKCLQDIVPGCNKVTGKAGMLDEIIN 192
Query: 206 YLKTLKLQLQVKFYVLQFSVTLFKLRFH 233
Y+++ LQ QV+F ++ SV +L H
Sbjct: 193 YVQS--LQQQVEFLSMKLSVINPELECH 218
>At2g24260 putative bHLH transcription factor (bHLH066)
Length = 350
Score = 67.4 bits (163), Expect = 6e-12
Identities = 30/51 (58%), Positives = 45/51 (87%)
Query: 166 HNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
H+++ER RR++I E+++AL+EL+PN NK DKASMLD+ IDY+K L+LQ++V
Sbjct: 149 HSIAERLRRERIAERMKALQELVPNGNKTDKASMLDEIIDYVKFLQLQVKV 199
>At5g58010 putative bHLH transcription factor (bHLH082)
Length = 274
Score = 66.2 bits (160), Expect = 1e-11
Identities = 34/78 (43%), Positives = 57/78 (72%), Gaps = 6/78 (7%)
Query: 140 TQENIVKEKPVREGNRVK-RSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKAS 198
T +V++KP RV+ R + H+++ER RR++I E++++L+EL+PN NK DKAS
Sbjct: 65 TSAPVVRQKP-----RVRARRGQATDPHSIAERLRRERIAERMKSLQELVPNTNKTDKAS 119
Query: 199 MLDDAIDYLKTLKLQLQV 216
MLD+ I+Y++ L+LQ++V
Sbjct: 120 MLDEIIEYVRFLQLQVKV 137
>At4g30980 putative bHLH transcription factor (bHLH069)
Length = 310
Score = 66.2 bits (160), Expect = 1e-11
Identities = 29/51 (56%), Positives = 45/51 (87%)
Query: 166 HNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
H+++ER RR++I E++++L+EL+PN NK DKASMLD+ IDY+K L+LQ++V
Sbjct: 141 HSIAERLRRERIAERMKSLQELVPNGNKTDKASMLDEIIDYVKFLQLQVKV 191
>At1g32640 transcription factor like protein, BHLH6
Length = 623
Score = 66.2 bits (160), Expect = 1e-11
Identities = 55/207 (26%), Positives = 91/207 (43%), Gaps = 23/207 (11%)
Query: 11 VNFSNFSIPAVFLKSNQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSGYE-RSRELLP 69
+ F N S + N KF LN +T S++ + RS E+L
Sbjct: 309 IQFENGSSSTITENPNLDPTPSPVHSQTQNPKFNNTFSRELNFSTSSSTLVKPRSGEILN 368
Query: 70 TVDEHSEAA-----ASHTNAPRIRGKRKASAINLCNAQKPSSVCSLEASNDLNFGVRKSH 124
DE ++ +S++ + KRK S + L L+FG + +
Sbjct: 369 FGDEGKRSSGNPDPSSYSGQTQFENKRKRSMV-------------LNEDKVLSFGDKTAG 415
Query: 125 EDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRAL 184
E S + +E V+++P + G + H +ER+RR+K+N++ AL
Sbjct: 416 E----SDHSDLEASVVKEVAVEKRPKKRGRKPANGREEPLNHVEAERQRREKLNQRFYAL 471
Query: 185 KELIPNCNKMDKASMLDDAIDYLKTLK 211
+ ++PN +KMDKAS+L DAI Y+ LK
Sbjct: 472 RAVVPNVSKMDKASLLGDAIAYINELK 498
>At4g02590 putative bHLH transcription factor (bHLH059)
Length = 310
Score = 65.1 bits (157), Expect = 3e-11
Identities = 27/51 (52%), Positives = 44/51 (85%)
Query: 166 HNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
H+++ER RR++I E+IRAL+EL+P NK D+A+M+D+ +DY+K L+LQ++V
Sbjct: 157 HSIAERLRRERIAERIRALQELVPTVNKTDRAAMIDEIVDYVKFLRLQVKV 207
>At1g03040 putative transcription factor (BHLH7)
Length = 302
Score = 63.9 bits (154), Expect = 7e-11
Identities = 26/51 (50%), Positives = 44/51 (85%)
Query: 166 HNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
H+++ER RR++I E+IR+L+EL+P NK D+A+M+D+ +DY+K L+LQ++V
Sbjct: 155 HSIAERLRRERIAERIRSLQELVPTVNKTDRAAMIDEIVDYVKFLRLQVKV 205
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,113,789
Number of Sequences: 26719
Number of extensions: 210648
Number of successful extensions: 1223
Number of sequences better than 10.0: 214
Number of HSP's better than 10.0 without gapping: 127
Number of HSP's successfully gapped in prelim test: 87
Number of HSP's that attempted gapping in prelim test: 1028
Number of HSP's gapped (non-prelim): 253
length of query: 240
length of database: 11,318,596
effective HSP length: 96
effective length of query: 144
effective length of database: 8,753,572
effective search space: 1260514368
effective search space used: 1260514368
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC139745.7


