

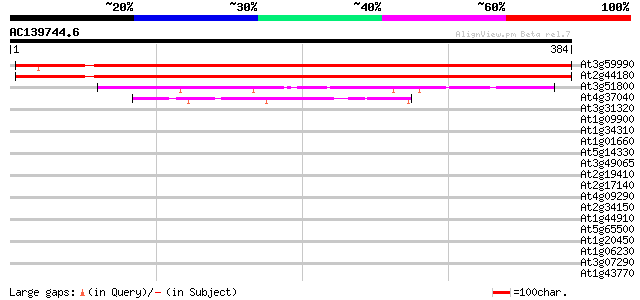
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139744.6 + phase: 1 /partial
(384 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g59990 unknown protein 545 e-155
At2g44180 methionine aminopeptidase-like protein 522 e-148
At3g51800 G2p (AtG2) 112 3e-25
At4g37040 methionine aminopeptidase like protein 50 3e-06
At3g31320 unknown protein 32 0.59
At1g09900 hypothetical protein 31 1.00
At1g34310 hypothetical protein 31 1.3
At1g01660 hypothetical protein 31 1.3
At5g14330 putative protein 30 1.7
At3g49065 unknown protein 30 1.7
At2g19410 putative protein kinase 30 2.2
At2g17140 hypothetical protein 30 2.2
At4g09290 putative protein 30 2.9
At2g34150 hypothetical protein 30 2.9
At1g44910 splicing factor like protein 29 3.8
At5g65500 putative protein 29 5.0
At1g20450 putative cold-acclimation protein 29 5.0
At1g06230 Ring3-like bromodomain protein 29 5.0
At3g07290 hypothetical protein 28 6.5
At1g43770 hypothetical protein 28 6.5
>At3g59990 unknown protein
Length = 439
Score = 545 bits (1403), Expect = e-155
Identities = 265/382 (69%), Positives = 314/382 (81%), Gaps = 7/382 (1%)
Query: 5 QKKKKNKKKKGKKV--QTDPPTIPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNEEK 62
+KKKKNK KK K++ QTDPP+IP+ ELF G +P G+I +Y N RT T+EEK
Sbjct: 62 KKKKKNKSKKKKELPQQTDPPSIPVVELFPSGEFPEGEIQEYKDDNLWRT-----TSEEK 116
Query: 63 RALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKA 122
R L+R E IYN VR AAE HRQ RK+++ +KPGM M ICE LENT RKLI E+GL+A
Sbjct: 117 RELERFEKPIYNSVRRAAEVHRQVRKYVRSIVKPGMLMTDICETLENTVRKLISENGLQA 176
Query: 123 GLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDK 182
G+AFPTGCS N AAH+TPN+GD TVL+YDDV K+DFGTHI+G IIDCAFT++FNP +D
Sbjct: 177 GIAFPTGCSLNWVAAHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIIDCAFTVAFNPMFDP 236
Query: 183 LIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISP 242
L+ A R+AT TGIK AGIDV LC+IGAAIQEVMESYEVE++GK +QVKSIRNLNGHSI P
Sbjct: 237 LLAASREATYTGIKEAGIDVRLCDIGAAIQEVMESYEVEINGKVFQVKSIRNLNGHSIGP 296
Query: 243 YRIHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLR 302
Y+IHAGK+VPIVKGGE T MEE E+YAIETFGSTG+G V +D++CSHYMKNFDAG++PLR
Sbjct: 297 YQIHAGKSVPIVKGGEQTKMEEGEFYAIETFGSTGKGYVREDLECSHYMKNFDAGHVPLR 356
Query: 303 LQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYT 362
L +K LL+ INKNFSTLAFC+R+LDR G TKY MALK+LCD GIV YPPLCDVKG Y
Sbjct: 357 LPRAKQLLATINKNFSTLAFCRRYLDRIGETKYLMALKNLCDSGIVQPYPPLCDVKGSYV 416
Query: 363 AQFEHTIMLRPTCKEVVSRGDD 384
+QFEHTI+LRPTCKEV+S+GDD
Sbjct: 417 SQFEHTILLRPTCKEVLSKGDD 438
>At2g44180 methionine aminopeptidase-like protein
Length = 441
Score = 522 bits (1344), Expect = e-148
Identities = 248/380 (65%), Positives = 306/380 (80%), Gaps = 5/380 (1%)
Query: 5 QKKKKNKKKKGKKVQTDPPTIPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNEEKRA 64
+KK K+KKKK QTDPP+IP+ ELF G +P G+I Y N RT T+EEKR
Sbjct: 66 KKKSKSKKKKSSLQQTDPPSIPVLELFPSGDFPQGEIQQYNDDNLWRT-----TSEEKRE 120
Query: 65 LDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGL 124
++R + IYN +R AAE HRQ RK+M+ +KPGM MI +CE LENT RKLI E+GL+AG+
Sbjct: 121 MERLQKPIYNSLRQAAEVHRQVRKYMRSILKPGMLMIDLCETLENTVRKLISENGLQAGI 180
Query: 125 AFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLI 184
AFPTGCS N+ AAH+TPN+GD TVL+YDDV K+DFGTHI+G I+D AFT++FNP +D L+
Sbjct: 181 AFPTGCSLNNVAAHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIVDSAFTVAFNPMFDPLL 240
Query: 185 EAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYR 244
A RDAT TGIK AG+DV LC++GAA+QEVMESYEVE++GK YQVKSIRNLNGHSI Y+
Sbjct: 241 AASRDATYTGIKEAGVDVRLCDVGAAVQEVMESYEVEINGKVYQVKSIRNLNGHSIGRYQ 300
Query: 245 IHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQ 304
IHA K+VP V+GGE T MEE E YAIETFGSTG+G V +D++CSHYMKN+D G++PLRL
Sbjct: 301 IHAEKSVPNVRGGEQTKMEEGELYAIETFGSTGKGYVREDLECSHYMKNYDVGHVPLRLP 360
Query: 305 SSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQ 364
+K LL+ INKNFSTLAFC+R+LDR G TKY MALK+LCD GI++ PP+CDVKG Y +Q
Sbjct: 361 RAKQLLATINKNFSTLAFCRRYLDRLGETKYLMALKNLCDSGIIEPCPPVCDVKGSYISQ 420
Query: 365 FEHTIMLRPTCKEVVSRGDD 384
FEHTI+LRPTCKE++S+GDD
Sbjct: 421 FEHTILLRPTCKEIISKGDD 440
>At3g51800 G2p (AtG2)
Length = 392
Score = 112 bits (281), Expect = 3e-25
Identities = 87/327 (26%), Positives = 157/327 (47%), Gaps = 24/327 (7%)
Query: 61 EKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLI----- 115
+++ L + ++ + + AAE + + + KP ++ ICE+ ++ ++
Sbjct: 8 DEKELSLTSPEVVTKYKSAAEIVNKALQVVLAECKPKAKIVDICEKGDSFIKEQTASMYK 67
Query: 116 -KEDGLKAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHING--RIIDCAF 172
+ ++ G+AFPT S N+ H++P A D +VLE D+ KID G HI+G ++
Sbjct: 68 NSKKKIERGVAFPTCISVNNTVGHFSPLASDESVLEDGDMVKIDMGCHIDGFIALVGHTH 127
Query: 173 TLSFNPKYDKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSI 232
L P + + + A NT AA + + L G +V E+ ++ Y K +
Sbjct: 128 VLQEGPLSGRKADVIA-AANT---AADVALRLVRPGKKNTDVTEA--IQKVAAAYDCKIV 181
Query: 233 RNLNGHSISPYRIHAGKTVPIVKGGEATV----MEENEYYAIETFGSTGRG--QVHDDMD 286
+ H + + I K V V E TV EENE YAI+ STG G ++ D+
Sbjct: 182 EGVLSHQLKQHVIDGNKVVLSVSSPETTVDEVEFEENEVYAIDIVASTGDGKPKLLDEKQ 241
Query: 287 CSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKG 346
+ Y K+ Y L++++S+ ++S I +NF + F R L+ + ++ L + + G
Sbjct: 242 TTIYKKDESVNYQ-LKMKASRFIISEIKQNFPRMPFTARSLEE---KRARLGLVECVNHG 297
Query: 347 IVDAYPPLCDVKGCYTAQFEHTIMLRP 373
+ YP L + G + AQ + T++L P
Sbjct: 298 HLQPYPVLYEKPGDFVAQIKFTVLLMP 324
>At4g37040 methionine aminopeptidase like protein
Length = 350
Score = 49.7 bits (117), Expect = 3e-06
Identities = 52/203 (25%), Positives = 83/203 (40%), Gaps = 29/203 (14%)
Query: 85 QTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLK------AGLAFPTGCSRNHCAAH 138
+ R + +KPG+T +I E + N +I E+G G S N C H
Sbjct: 125 RVRDYAGTLVKPGVTTDEIDEAVHN----MIIENGAYPSPLGYGGFPKSVCTSVNECICH 180
Query: 139 YTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTL---SFNPKYDKLIEAVRDATNTGI 195
P D LE D+ ID ++NG D + T + + K KL+E +++ + I
Sbjct: 181 GIP---DSRPLEDGDIINIDVTVYLNGYHGDTSATFFCGNVDEKAKKLVEVTKESLDKAI 237
Query: 196 KAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVPIVK 255
G V +IG I ++ + ++ + +R GH + HA V +
Sbjct: 238 SICGPGVEYKKIGKVIHDLADKHKYGV---------VRQFVGHGVGSV-FHADPVVLHFR 287
Query: 256 GGEATVMEENEYYAIE---TFGS 275
EA M N+ + IE T GS
Sbjct: 288 NNEAGRMVLNQTFTIEPMLTIGS 310
>At3g31320 unknown protein
Length = 327
Score = 32.0 bits (71), Expect = 0.58
Identities = 14/26 (53%), Positives = 17/26 (64%)
Query: 3 EAQKKKKNKKKKGKKVQTDPPTIPIC 28
+A K KK KKKK K+V DPP +C
Sbjct: 89 KADKGKKKKKKKKKQVMPDPPGSTLC 114
>At1g09900 hypothetical protein
Length = 598
Score = 31.2 bits (69), Expect = 1.00
Identities = 29/121 (23%), Positives = 48/121 (38%), Gaps = 22/121 (18%)
Query: 279 GQVHDDMDCSHYMKNF-------DAGYMPLRLQSSKSLLSVINKNFSTLAFCKRW----- 326
G V D + C+ ++ F A + L+ S ++ VI N +CK
Sbjct: 132 GNVPDIIPCTTLIRGFCRLGKTRKAAKILEILEGSGAVPDVITYNVMISGYCKAGEINNA 191
Query: 327 ---LDRAGCTK----YQMALKDLCDKGIVDAYPPLCD---VKGCYTAQFEHTIMLRPTCK 376
LDR + Y L+ LCD G + + D + CY +TI++ TC+
Sbjct: 192 LSVLDRMSVSPDVVTYNTILRSLCDSGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCR 251
Query: 377 E 377
+
Sbjct: 252 D 252
>At1g34310 hypothetical protein
Length = 591
Score = 30.8 bits (68), Expect = 1.3
Identities = 11/25 (44%), Positives = 16/25 (64%)
Query: 170 CAFTLSFNPKYDKLIEAVRDATNTG 194
C FT+ + P YDK ++AV + N G
Sbjct: 265 CMFTVVYKPSYDKFLDAVNNKFNVG 289
>At1g01660 hypothetical protein
Length = 568
Score = 30.8 bits (68), Expect = 1.3
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 2/33 (6%)
Query: 212 QEVMESYEVELDGKTYQVKSIRNL--NGHSISP 242
QEVM V DG TY+ +S+R NGH SP
Sbjct: 507 QEVMREPRVAADGFTYEAESLREWLDNGHETSP 539
>At5g14330 putative protein
Length = 128
Score = 30.4 bits (67), Expect = 1.7
Identities = 13/21 (61%), Positives = 15/21 (70%)
Query: 2 GEAQKKKKNKKKKGKKVQTDP 22
GE Q+KK NKKKK K +Q P
Sbjct: 38 GEQQRKKPNKKKKEKNIQMAP 58
>At3g49065 unknown protein
Length = 805
Score = 30.4 bits (67), Expect = 1.7
Identities = 18/41 (43%), Positives = 23/41 (55%), Gaps = 5/41 (12%)
Query: 204 LCEIGAAIQEVMESYEVELDGKTYQVKSIRN--LNGHSISP 242
LC I QEVM+ + DG TY+ ++IR NGH SP
Sbjct: 740 LCPI---FQEVMKDPLIAADGFTYEAEAIREWLANGHDTSP 777
>At2g19410 putative protein kinase
Length = 801
Score = 30.0 bits (66), Expect = 2.2
Identities = 19/63 (30%), Positives = 35/63 (55%), Gaps = 3/63 (4%)
Query: 183 LIEAVRDATNTGIKAAGIDV--PLCEIGAAIQEVMESYEVELDGKTYQVKSIRN-LNGHS 239
+++ + + N+ +K G ++ P ++E+ME E+ DG TY+ K+I L H+
Sbjct: 704 VLKRLVETANSKVKKEGSNLRAPSHYFCPILREIMEEPEIAADGFTYERKAILAWLEKHN 763
Query: 240 ISP 242
ISP
Sbjct: 764 ISP 766
>At2g17140 hypothetical protein
Length = 903
Score = 30.0 bits (66), Expect = 2.2
Identities = 16/45 (35%), Positives = 23/45 (50%), Gaps = 3/45 (6%)
Query: 335 YQMALKDLCDKGIVDAYPPLCD---VKGCYTAQFEHTIMLRPTCK 376
+ + ++ LCD VDA L D KGC +F I++R CK
Sbjct: 150 FNLLIRALCDSSCVDAARELFDEMPEKGCKPNEFTFGILVRGYCK 194
>At4g09290 putative protein
Length = 376
Score = 29.6 bits (65), Expect = 2.9
Identities = 12/14 (85%), Positives = 14/14 (99%)
Query: 1 GGEAQKKKKNKKKK 14
GG+AQKKKKNKKK+
Sbjct: 80 GGDAQKKKKNKKKE 93
>At2g34150 hypothetical protein
Length = 604
Score = 29.6 bits (65), Expect = 2.9
Identities = 39/131 (29%), Positives = 62/131 (46%), Gaps = 17/131 (12%)
Query: 151 YDDVTKIDFGTHINGRIID---CAFT----LSFNPKY--DKLIE-----AVRDATNTGIK 196
Y+D+T+ + N +D CA T LS +P Y D+LI A ++ T
Sbjct: 239 YNDITEQETEKISNNFSVDETKCAATSELHLSSSPVYKSDELIHQDPWAASEISSGTHSY 298
Query: 197 AAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIH-AGKTVP-IV 254
+ G PL +I + IQE ES EVE T +K+ N N + P + +T+P IV
Sbjct: 299 SNGFSNPLYDI-SGIQEHQESEEVESSCDTESIKTWTNGNLLGLKPSKPKIIAETIPEIV 357
Query: 255 KGGEATVMEEN 265
+ ++ +E+
Sbjct: 358 EDIDSETFQEH 368
>At1g44910 splicing factor like protein
Length = 958
Score = 29.3 bits (64), Expect = 3.8
Identities = 26/122 (21%), Positives = 51/122 (41%), Gaps = 16/122 (13%)
Query: 3 EAQKKKKNKKKKGKKVQTDPPT-IPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNEE 61
E +++K+ K++ K+ ++D T + + E D + R KDR
Sbjct: 841 EREREKEKGKERSKREESDGETAMDVSEGHKD---------------EKRKGKDRDRKHR 885
Query: 62 KRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLK 121
+R + S+ D+ ++ E+ + +RKH K E EN ++ KE +
Sbjct: 886 RRHHNNSDEDVSSDRDDRDESKKSSRKHGNDRKKSRKHANSPESESENRHKRQKKESSRR 945
Query: 122 AG 123
+G
Sbjct: 946 SG 947
>At5g65500 putative protein
Length = 765
Score = 28.9 bits (63), Expect = 5.0
Identities = 18/57 (31%), Positives = 32/57 (55%), Gaps = 3/57 (5%)
Query: 189 DATNTGIKAAGI-DVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRN--LNGHSISP 242
+ATN+ + D+P + +QEVM++ V DG +Y++++I+ GH SP
Sbjct: 675 EATNSNMDEGDPNDIPSVFMCPILQEVMKNPHVAADGFSYELEAIQEWLSMGHDTSP 731
>At1g20450 putative cold-acclimation protein
Length = 260
Score = 28.9 bits (63), Expect = 5.0
Identities = 19/73 (26%), Positives = 31/73 (42%)
Query: 183 LIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISP 242
+++ +++ G K G DVP+ A V + E + K + K L GHS P
Sbjct: 146 VMDRIKEKFPLGEKPGGDDVPVVTTMPAPHSVEDHKPEEEEKKGFMDKIKEKLPGHSKKP 205
Query: 243 YRIHAGKTVPIVK 255
T P+V+
Sbjct: 206 EDSQVVNTTPLVE 218
>At1g06230 Ring3-like bromodomain protein
Length = 766
Score = 28.9 bits (63), Expect = 5.0
Identities = 16/61 (26%), Positives = 28/61 (45%), Gaps = 4/61 (6%)
Query: 55 DRFTNEEKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKL 114
DRF K+ L + + + LA +A + ++ QQ + P + E NTA+K
Sbjct: 664 DRFVTNYKKGLSKKK----RKAELAIQARAEAERNSQQQMAPAPAAHEFSREGGNTAKKT 719
Query: 115 I 115
+
Sbjct: 720 L 720
>At3g07290 hypothetical protein
Length = 880
Score = 28.5 bits (62), Expect = 6.5
Identities = 13/46 (28%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query: 335 YQMALKDLCDKGIVDAYPPLCD---VKGCYTAQFEHTIMLRPTCKE 377
Y + +K LCD+G++D L D +GC +T+++ C++
Sbjct: 304 YTVLIKALCDRGLIDKAFNLFDEMIPRGCKPNVHTYTVLIDGLCRD 349
>At1g43770 hypothetical protein
Length = 431
Score = 28.5 bits (62), Expect = 6.5
Identities = 11/25 (44%), Positives = 15/25 (60%)
Query: 3 EAQKKKKNKKKKGKKVQTDPPTIPI 27
E +KKK KKKK K + PP + +
Sbjct: 156 ERSEKKKKKKKKKKSINHSPPVLAV 180
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,989,311
Number of Sequences: 26719
Number of extensions: 381438
Number of successful extensions: 1220
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1197
Number of HSP's gapped (non-prelim): 28
length of query: 384
length of database: 11,318,596
effective HSP length: 101
effective length of query: 283
effective length of database: 8,619,977
effective search space: 2439453491
effective search space used: 2439453491
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC139744.6


