

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139744.2 - phase: 0 /pseudo
(1366 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
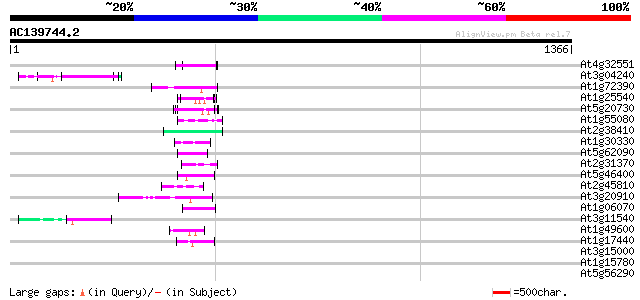
Score E
Sequences producing significant alignments: (bits) Value
At4g32551 Leunig protein 67 7e-11
At3g04240 O-linked GlcNAc transferase like protein 62 2e-09
At1g72390 unknown protein 57 1e-07
At1g25540 hypothetical protein 56 1e-07
At5g20730 auxin response factor 7 (ARF7) 55 2e-07
At1g55080 unknown protein 55 2e-07
At2g38410 unknown protein 53 1e-06
At1g30330 putative protein 52 2e-06
At5g62090 unknown protein 50 9e-06
At2g31370 bZIP transcription factor PosF21 / AtbZip59 50 1e-05
At5g46400 putative protein 48 4e-05
At2g45810 putative ATP-dependent RNA helicase 47 8e-05
At3g20910 CCAAT-binding factor B chain like protein 47 1e-04
At1g06070 bZip transcription factor AtbZip69 45 3e-04
At3g11540 spindly (gibberellin signal transduction protein) 44 6e-04
At1g49600 DNA binding protein ACBF, putative 44 8e-04
At1g17440 unknown protein 44 8e-04
At3g15000 unknown protein 43 0.001
At1g15780 unknown protein 43 0.001
At5g56290 peroxisomal targeting signal type 1 receptor 42 0.002
>At4g32551 Leunig protein
Length = 931
Score = 67.0 bits (162), Expect = 7e-11
Identities = 43/100 (43%), Positives = 51/100 (51%), Gaps = 3/100 (3%)
Query: 405 ARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQ-HHYRLMQQHQQQL 463
AR Q Q Q P QQQ Q Q + QQ+QQ QQ Q HH++ QQ QQQ
Sbjct: 86 AREQQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQ 145
Query: 464 RMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCP 503
+ QQQQQQQ H P QQ Q + P Q QQP+ + P
Sbjct: 146 QQQQQQQQQQHQNQPPSQQQQQQSTPQHQ--QQPTPQQQP 183
Score = 45.8 bits (107), Expect = 2e-04
Identities = 34/84 (40%), Positives = 40/84 (47%), Gaps = 8/84 (9%)
Query: 422 QQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQ 481
+QQLQ Q H + QQ+QQ QQ Q L+Q+ QQQ QQQQQQ H Q
Sbjct: 88 EQQLQQSQ----HPQVSQQQQQQQQQQIQMQQLLLQRAQQQ----QQQQQQQHHHHQQQQ 139
Query: 482 QPGQVARPPQQSLQQPSVKSCPPQ 505
Q Q + QQ QQ P Q
Sbjct: 140 QQQQQQQQQQQQQQQQHQNQPPSQ 163
Score = 42.7 bits (99), Expect = 0.001
Identities = 31/86 (36%), Positives = 40/86 (46%), Gaps = 2/86 (2%)
Query: 419 YLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVS 478
Y+ Q ++ + Q +S +PQ QQ QQ Q MQQ Q QQQQQQQ
Sbjct: 78 YIETQMIKARE--QQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHH 135
Query: 479 PVQQPGQVARPPQQSLQQPSVKSCPP 504
QQ Q + QQ QQ ++ PP
Sbjct: 136 QQQQQQQQQQQQQQQQQQQQHQNQPP 161
Score = 38.5 bits (88), Expect = 0.027
Identities = 23/67 (34%), Positives = 31/67 (45%)
Query: 406 RPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM 465
R Q Q QQ ++ +QQQ Q Q Q + Q + Q Q + QHQQQ
Sbjct: 120 RAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTP 179
Query: 466 QQQQQQQ 472
QQQ Q++
Sbjct: 180 QQQPQRR 186
>At3g04240 O-linked GlcNAc transferase like protein
Length = 977
Score = 62.4 bits (150), Expect = 2e-09
Identities = 54/232 (23%), Positives = 102/232 (43%), Gaps = 18/232 (7%)
Query: 21 RAVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEI 80
RA++ +++ + + P F A +L LG ++K P A+ + Y A
Sbjct: 241 RALQYYKEAVKLKPAFPDA---YLNLGNVYKALGRPTEAI----MCYQHALQMRPNSAMA 293
Query: 81 QFHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMT 140
+IA +Y Q + LA Y+Q L P L+A Y+N+ R+
Sbjct: 294 FGNIASIYYEQGQLDLAIRHYKQALSRD--PRFLEA---------YNNLGNALKDIGRVD 342
Query: 141 LAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVL 200
A+RC + L N Q + LG Y A ++ + TT + + ++ ++
Sbjct: 343 EAVRCYNQCLALQPNHPQAMANLGNIYMEWNMMGPASSLFKATLAVTTGLSAPFNNLAII 402
Query: 201 YQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIHSV 252
Y+QQ Y DA+ Y +++D + + +N G Y+ ++ +A+ Y+H++
Sbjct: 403 YKQQGNYSDAISCYNEVLRIDPLAADALVNRGNTYKEIGRVTEAIQDYMHAI 454
Score = 49.3 bits (116), Expect = 2e-05
Identities = 31/139 (22%), Positives = 63/139 (45%)
Query: 126 YHNVEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVE 185
+ N+ + + R++ A +C +++L + LG + G A+ Y V
Sbjct: 158 WSNLASAYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIHEAYSCYLEAVR 217
Query: 186 KTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDAL 245
A W ++ L+ + ALQ Y AV+L +++NLG +Y++ + +A+
Sbjct: 218 IQPTFAIAWSNLAGLFMESGDLNRALQYYKEAVKLKPAFPDAYLNLGNVYKALGRPTEAI 277
Query: 246 ACYIHSVRASKDKAAGVGS 264
CY H+++ + A G+
Sbjct: 278 MCYQHALQMRPNSAMAFGN 296
Score = 45.8 bits (107), Expect = 2e-04
Identities = 43/194 (22%), Positives = 81/194 (41%), Gaps = 15/194 (7%)
Query: 68 VDASPSTFCKLEIQFHIAHLYEVQKKHKLAKELYE-----QLLKEKDLPLH---LKADIY 119
+ S S+ L QF+ H + + LA +LY+ Q L+ ++ L+ D
Sbjct: 31 LSVSSSSSSSLLQQFNKTHEGDDDARLALAHQLYKGGDFKQALEHSNMVYQRNPLRTDNL 90
Query: 120 RQLGWMYHNVEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDA 179
+G +Y+ ++ + R A+R + E N + + G D A
Sbjct: 91 LLIGAIYYQLQEYDMCIARNEEALRIQPQFAECYGN-------MANAWKEKGDTDRAIRY 143
Query: 180 YRNVVEKTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCS 239
Y +E AD W ++ Y ++ + +A Q A+ L+ + NLG L ++
Sbjct: 144 YLIAIELRPNFADAWSNLASAYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQG 203
Query: 240 QLKDALACYIHSVR 253
+ +A +CY+ +VR
Sbjct: 204 LIHEAYSCYLEAVR 217
Score = 45.8 bits (107), Expect = 2e-04
Identities = 35/148 (23%), Positives = 60/148 (39%), Gaps = 1/148 (0%)
Query: 126 YHNVEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVE 185
+ N+ + + ++ LAIR +++L D + LG IG+ D A Y +
Sbjct: 294 FGNIASIYYEQGQLDLAIRHYKQALSRDPRFLEAYNNLGNALKDIGRVDEAVRCYNQCLA 353
Query: 186 KTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDAL 245
++G +Y + A + + + SA + NL I+Y+ DA+
Sbjct: 354 LQPNHPQAMANLGNIYMEWNMMGPASSLFKATLAVTTGLSAPFNNLAIIYKQQGNYSDAI 413
Query: 246 ACYIHSVRASKDKA-AGVGSGTTSSESG 272
+CY +R A A V G T E G
Sbjct: 414 SCYNEVLRIDPLAADALVNRGNTYKEIG 441
>At1g72390 unknown protein
Length = 1088
Score = 56.6 bits (135), Expect = 1e-07
Identities = 56/172 (32%), Positives = 72/172 (41%), Gaps = 26/172 (15%)
Query: 346 GMAQQGGPK--PNHYVNGVNGGAVPPPPYPGA--VPGAGPNAKRFKPNGEVDNGMVGAAG 401
GMA +GG P ++GV+G P G + N +R AA
Sbjct: 857 GMANRGGVMGAPQTGISGVSGTRQMHPSSAGLSMLDQNRANLQR-------------AAA 903
Query: 402 MGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQ 461
MG+ PP+ P Y+NQQQ Q Q + L QQ+ Q Q +L Q QQ
Sbjct: 904 MGNMGPPKLMPGM-MNLYMNQQQQQQQLQQQPQQQQLQHQQQLQQPMSQPSQQLAQSPQQ 962
Query: 462 QLRMQ-----QQQQQQLHAGVSPVQ---QPGQVARPPQQSLQQPSVKSCPPQ 505
Q ++Q QQ QQQ A SP+Q P QV P QQ +S P Q
Sbjct: 963 QQQLQQHEQPQQAQQQQQATASPLQSVLSPPQVGSPSAGITQQQLQQSSPQQ 1014
>At1g25540 hypothetical protein
Length = 809
Score = 56.2 bits (134), Expect = 1e-07
Identities = 40/101 (39%), Positives = 45/101 (43%), Gaps = 19/101 (18%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ +L QQQ+ LQ Q QQ QQ QQ QH +Q HQQQ + QQQ
Sbjct: 671 QIQQQQQQQQHLQQQQMPQLQQQQ--------QQHQQQQQQQHQLSQLQHHQQQQQQQQQ 722
Query: 469 QQQ-----------QLHAGVSPVQQPGQVARPPQQSLQQPS 498
QQQ Q SP+ Q Q P Q QQ S
Sbjct: 723 QQQQHQLTQLQHHHQQQQQASPLNQMQQQTSPLNQMQQQTS 763
Score = 50.1 bits (118), Expect = 9e-06
Identities = 35/101 (34%), Positives = 48/101 (46%), Gaps = 5/101 (4%)
Query: 408 PQAQPQQPPPYYLNQQQLQL--LQYYQSHVESLNPQQRQ-QMQQFQHHYRLMQQHQQQLR 464
PQ Q QQ QQQ QL LQ++Q + QQ+Q Q+ Q QHH++ QQ +
Sbjct: 688 PQLQQQQQQHQQQQQQQHQLSQLQHHQQQQQQQQQQQQQHQLTQLQHHHQQQQQASPLNQ 747
Query: 465 MQQQQQ--QQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCP 503
MQQQ Q+ SP+ Q Q +P Q + + P
Sbjct: 748 MQQQTSPLNQMQQQTSPLNQMQQQQQPQQMVMGGQAFAQAP 788
Score = 48.9 bits (115), Expect = 2e-05
Identities = 32/80 (40%), Positives = 41/80 (51%), Gaps = 2/80 (2%)
Query: 417 PYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAG 476
P NQQQ Q Q +Q + QQ+QQ QQ ++ Q QQQ + QQQQQQQ
Sbjct: 649 PQIPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQ-- 706
Query: 477 VSPVQQPGQVARPPQQSLQQ 496
+S +Q Q + QQ QQ
Sbjct: 707 LSQLQHHQQQQQQQQQQQQQ 726
Score = 48.5 bits (114), Expect = 3e-05
Identities = 40/105 (38%), Positives = 47/105 (44%), Gaps = 17/105 (16%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQF-----QHHYRLMQQHQ-- 460
P Q QQ + QQQ Q +Q Q + L Q+QQM Q QH + QQHQ
Sbjct: 652 PNQQQQQQQQLHQQQQQQQQIQQQQQQQQHL---QQQQMPQLQQQQQQHQQQQQQQHQLS 708
Query: 461 -----QQLRMQQQQQQQLH--AGVSPVQQPGQVARPPQQSLQQPS 498
QQ + QQQQQQQ H + Q Q A P Q QQ S
Sbjct: 709 QLQHHQQQQQQQQQQQQQHQLTQLQHHHQQQQQASPLNQMQQQTS 753
Score = 38.1 bits (87), Expect = 0.035
Identities = 23/65 (35%), Positives = 31/65 (47%), Gaps = 7/65 (10%)
Query: 431 YQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPP 490
++ + + QQ+QQ+ Q QQ QQQ++ QQQQQQ L P Q Q
Sbjct: 647 FKPQIPNQQQQQQQQLHQ-------QQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQ 699
Query: 491 QQSLQ 495
QQ Q
Sbjct: 700 QQQQQ 704
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 55.5 bits (132), Expect = 2e-07
Identities = 41/108 (37%), Positives = 52/108 (47%), Gaps = 14/108 (12%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQ 467
P +Q QPP QQQLQ L + SLN QQ+Q Q Q + + Q QQQL+ QQ
Sbjct: 488 PMSQLPQPPTTLSQQQQLQQLLH-----SSLNHQQQQSQSQQQQQQQQLLQQQQQLQSQQ 542
Query: 468 ---------QQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQA 506
QQQQQL Q Q +P QQ QQ +++ P Q+
Sbjct: 543 HSNNNQSQSQQQQQLLQQQQQQQLQQQHQQPLQQQTQQQQLRTQPLQS 590
Score = 54.3 bits (129), Expect = 5e-07
Identities = 39/103 (37%), Positives = 50/103 (47%), Gaps = 8/103 (7%)
Query: 400 AGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH 459
+ + H + QQ L QQQ QL S+ QQ+QQ+ Q Q +L QQH
Sbjct: 511 SSLNHQQQQSQSQQQQQQQQLLQQQQQLQSQQHSNNNQSQSQQQQQLLQQQQQQQLQQQH 570
Query: 460 QQQLRMQQQQQQQLHAGVSPVQ-----QPGQVARPPQQSLQQP 497
QQ L+ QQ QQQQL P+Q QP Q+ + Q LQ P
Sbjct: 571 QQPLQ-QQTQQQQLR--TQPLQSHSHPQPQQLQQHKLQQLQVP 610
Score = 47.0 bits (110), Expect = 8e-05
Identities = 37/106 (34%), Positives = 47/106 (43%), Gaps = 11/106 (10%)
Query: 404 HARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQL 463
H+ Q+Q QQ QQQ QL Q +Q ++ QQ+ + Q Q H Q QQ
Sbjct: 543 HSNNNQSQSQQQQQLLQQQQQQQLQQQHQQPLQQQTQQQQLRTQPLQSHSHPQPQQLQQH 602
Query: 464 RMQQQQ--QQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAV 507
++QQ Q Q QL+ G QQ Q Q QQ S PQ V
Sbjct: 603 KLQQLQVPQNQLYNG----QQAAQ-----QHQSQQASTHHLQPQLV 639
Score = 40.0 bits (92), Expect = 0.009
Identities = 37/116 (31%), Positives = 48/116 (40%), Gaps = 20/116 (17%)
Query: 409 QAQPQQPPPYY-LNQQQLQLLQYYQSHVESLNPQQ--RQQMQQFQHHYRLMQQHQQQL-- 463
Q + QQPP NQQ L + +Q+ + + Q Q QFQ RL QQ QQQ
Sbjct: 708 QVEQQQPPGLNGQNQQTLLQQKAHQAQAQQIFQQSLLEQPHIQFQLLQRLQQQQQQQFLS 767
Query: 464 --------RMQQQQQQQLHAGVSPVQQPG-------QVARPPQQSLQQPSVKSCPP 504
++Q QQ QQL Q P +PPQ + +P K PP
Sbjct: 768 PQSQLPHHQLQSQQLQQLPTLSQGHQFPSSCTNNGLSTLQPPQMLVSRPQEKQNPP 823
>At1g55080 unknown protein
Length = 244
Score = 55.5 bits (132), Expect = 2e-07
Identities = 44/111 (39%), Positives = 51/111 (45%), Gaps = 25/111 (22%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ ++L QQQ Q + PQQ+Q+MQQ+Q Q QQ QQQ
Sbjct: 43 QQQQQQQQQFHLQQQQQT-----QQQQQQFQPQQQQEMQQYQQF-----QQQQHFIQQQQ 92
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQ--AVHIPFSFLSTP 517
QQQ SP QP QSLQ P PPQ VH P S + TP
Sbjct: 93 FQQQQRLLQSPPLQP--------QSLQSP-----PPQQTMVHTPQSMMHTP 130
Score = 37.7 bits (86), Expect = 0.046
Identities = 33/94 (35%), Positives = 43/94 (45%), Gaps = 10/94 (10%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQ---HQQQLRM 465
Q Q QQ + QQQ Q +Q YQ + Q Q QQFQ RL+Q Q L+
Sbjct: 57 QQQTQQQQQQFQPQQQ-QEMQQYQQFQQQ---QHFIQQQQFQQQQRLLQSPPLQPQSLQS 112
Query: 466 QQQQQQQLHAGVSPV---QQPGQVARPPQQSLQQ 496
QQ +H S + QQ Q+ + P Q+ QQ
Sbjct: 113 PPPQQTMVHTPQSMMHTPQQQQQLVQTPVQTPQQ 146
>At2g38410 unknown protein
Length = 671
Score = 52.8 bits (125), Expect = 1e-06
Identities = 43/147 (29%), Positives = 57/147 (38%), Gaps = 4/147 (2%)
Query: 375 AVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQL---QLLQYY 431
A+P P K +D + P +QP PPP +Q Q +
Sbjct: 416 ALPDPPPPVNTTKEQDMIDLLSITLCTPSTPPAPSSQPSPPPPAGSDQNTHIYPQPQPRF 475
Query: 432 QSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQ 491
S+V QQ+ Q Q Q Y QQHQQQ Q Q Q G S +QQP Q +
Sbjct: 476 DSYVAPWAQQQQPQQPQAQQGYSQHQQHQQQQGYSQPQHSQQQQGYSQLQQP-QPQQGYS 534
Query: 492 QSLQQPSVKSCPPQAVHIPFSFLSTPF 518
QS Q V+ P P+ + P+
Sbjct: 535 QSQPQAQVQMQPSTRPQNPYEYPPPPW 561
>At1g30330 putative protein
Length = 933
Score = 52.0 bits (123), Expect = 2e-06
Identities = 38/88 (43%), Positives = 46/88 (52%), Gaps = 5/88 (5%)
Query: 401 GMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQ 460
G P QPQ L+QQQ QL Q Q + L+ QQ+QQ+ Q Q +L QQ Q
Sbjct: 468 GFSMQSPSLVQPQMLQQQ-LSQQQQQLSQQQQQQ-QQLSQQQQQQLSQ-QQQQQLSQQQQ 524
Query: 461 QQLRMQQQQQQQLHAGVSPVQQPGQVAR 488
QQL QQQQQQ + GV QP A+
Sbjct: 525 QQL--SQQQQQQAYLGVPETHQPQSQAQ 550
Score = 42.0 bits (97), Expect = 0.002
Identities = 33/93 (35%), Positives = 42/93 (44%), Gaps = 5/93 (5%)
Query: 411 QPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQ 470
Q Q P + + L Q Q + L+ QQ+Q QQ Q +L QQ QQQL QQQQQ
Sbjct: 462 QFQNSPGFSMQSPSLVQPQMLQ---QQLSQQQQQLSQQQQQQQQLSQQQQQQLSQQQQQQ 518
Query: 471 --QQLHAGVSPVQQPGQVARPPQQSLQQPSVKS 501
QQ +S QQ P+ Q +S
Sbjct: 519 LSQQQQQQLSQQQQQQAYLGVPETHQPQSQAQS 551
Score = 40.0 bits (92), Expect = 0.009
Identities = 44/158 (27%), Positives = 60/158 (37%), Gaps = 24/158 (15%)
Query: 410 AQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQ--- 466
+Q QQ QQQ QL Q Q + QQ Q QQ Q + + + HQ Q + Q
Sbjct: 494 SQQQQQQQQLSQQQQQQLSQQQQQQLSQQQQQQLSQQQQQQAYLGVPETHQPQSQAQSQS 553
Query: 467 ----QQQQQQL----------HAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAV----- 507
QQQQQ+ A VS + Q G ++P LQ C Q+
Sbjct: 554 NNHLSQQQQQVVDNHNPSASSAAVVSAMSQFGSASQPNTSPLQS-MTSLCHQQSFSDTNG 612
Query: 508 -HIPFSFLSTPFFDMTTDRS*YIMIKCSFTDRSCSSVW 544
+ P S L T + + D S ++ SS W
Sbjct: 613 GNNPISPLHTLLSNFSQDESSQLLHLTRTNSAMTSSGW 650
Score = 33.5 bits (75), Expect = 0.86
Identities = 23/70 (32%), Positives = 29/70 (40%)
Query: 427 LLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQV 486
LLQ+ S S+ Q Q Q QQ Q + QQQQ Q QQ Q+
Sbjct: 460 LLQFQNSPGFSMQSPSLVQPQMLQQQLSQQQQQLSQQQQQQQQLSQQQQQQLSQQQQQQL 519
Query: 487 ARPPQQSLQQ 496
++ QQ L Q
Sbjct: 520 SQQQQQQLSQ 529
>At5g62090 unknown protein
Length = 816
Score = 50.1 bits (118), Expect = 9e-06
Identities = 34/78 (43%), Positives = 43/78 (54%), Gaps = 6/78 (7%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQR---QQMQQFQHHYRLMQQHQQQLRM 465
Q Q Q P + + QQ Q L+ Q +++SL P QR QQ QQ Q +L QQHQQQ +
Sbjct: 212 QQQQGQNPQFQILLQQ-QKLRQQQQYLQSLPPLQRVQLQQQQQVQQQQQLQQQHQQQQQQ 270
Query: 466 QQQQ--QQQLHAGVSPVQ 481
QQQ Q QL G P +
Sbjct: 271 LQQQGMQMQLTGGPRPYE 288
Score = 35.8 bits (81), Expect = 0.17
Identities = 33/90 (36%), Positives = 43/90 (47%), Gaps = 17/90 (18%)
Query: 424 QLQLLQYYQSHVESLNPQQRQQMQ----QFQ-----HHYRLMQQHQQQL----RMQQQQQ 470
Q Q+L+ + + L QQ+QQ Q QFQ R QQ+ Q L R+Q QQQ
Sbjct: 192 QQQILRQWLQRQDILQQQQQQQQQGQNPQFQILLQQQKLRQQQQYLQSLPPLQRVQLQQQ 251
Query: 471 QQLHAGVSPVQQPGQVARPPQQSLQQPSVK 500
QQ V QQ Q + QQ LQQ ++
Sbjct: 252 QQ----VQQQQQLQQQHQQQQQQLQQQGMQ 277
>At2g31370 bZIP transcription factor PosF21 / AtbZip59
Length = 398
Score = 49.7 bits (117), Expect = 1e-05
Identities = 35/93 (37%), Positives = 46/93 (48%), Gaps = 18/93 (19%)
Query: 418 YYLNQQQLQLL----QYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQL 473
+Y N Q +Q + Q+ Q + S QQ+QQ QQ QH + QQ QQQ + QQQQ QQL
Sbjct: 316 FYSNNQSMQTILAAKQFQQLQIHSQKQQQQQQQQQQQHQQQ--QQQQQQYQFQQQQMQQL 373
Query: 474 HAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQA 506
+ + QQ QQ V+ P QA
Sbjct: 374 ------------MQQRLQQQEQQNGVRLKPSQA 394
Score = 34.7 bits (78), Expect = 0.39
Identities = 32/98 (32%), Positives = 45/98 (45%), Gaps = 16/98 (16%)
Query: 380 GPNAKRFKPNGEVDNGMVGAAGMG----HARPPQAQPQQPPPYYLNQQQLQLLQYYQSHV 435
G N ++F N + ++ A H++ Q Q QQ + QQQ Q Q YQ
Sbjct: 310 GSNQQQFYSNNQSMQTILAAKQFQQLQIHSQKQQQQQQQQQQQHQQQQQQQ--QQYQF-- 365
Query: 436 ESLNPQQRQQMQQFQHHYRLMQQHQQQ-LRMQQQQQQQ 472
Q+QQMQQ RL QQ QQ +R++ Q Q+
Sbjct: 366 ------QQQQMQQLMQQ-RLQQQEQQNGVRLKPSQAQK 396
>At5g46400 putative protein
Length = 1022
Score = 47.8 bits (112), Expect = 4e-05
Identities = 39/102 (38%), Positives = 47/102 (45%), Gaps = 13/102 (12%)
Query: 409 QAQPQQPPPYYLNQQQLQLL----------QYYQ-SHVESLNPQQRQQMQQFQHHYRLMQ 457
Q QP Q P L+Q +QLL QY Q VE +N QQ+ Q Q Q + Q
Sbjct: 903 QPQPNQNPQPQLDQNLVQLLSKQYQSQAKTQYLQPQQVEQVNTQQQSQEPQNQQQIQFQQ 962
Query: 458 QHQQQLRMQQQQ--QQQLHAGVSPVQQPGQVARPPQQSLQQP 497
Q QQQ QQQQ QQQ + QQ A+ +Q L P
Sbjct: 963 QQQQQEWFQQQQQWQQQQYLLYIQQQQLQGEAKGDEQRLSMP 1004
Score = 33.1 bits (74), Expect = 1.1
Identities = 27/82 (32%), Positives = 38/82 (45%), Gaps = 6/82 (7%)
Query: 407 PPQAQ-PQQPPPYYLNQQQLQLL----QYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQ 461
PP++Q PQ +Q Q Q Q+ V+S N QQ QMQ + + ++ QQ+
Sbjct: 830 PPESQNPQNQCQNSTSQVQTSFAYPQTQIPQNPVQS-NYQQEGQMQSHEAYNQMWQQYYY 888
Query: 462 QLRMQQQQQQQLHAGVSPVQQP 483
QQQQQ + P Q P
Sbjct: 889 SYYYYQQQQQLMSEQPQPNQNP 910
>At2g45810 putative ATP-dependent RNA helicase
Length = 528
Score = 47.0 bits (110), Expect = 8e-05
Identities = 37/104 (35%), Positives = 44/104 (41%), Gaps = 19/104 (18%)
Query: 369 PPPYPGAVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLL 428
PP A PG PN + PN + + P QPQ PP YL Q
Sbjct: 12 PPGIGAAGPGPDPNFQSRNPNPPQPQQYLQSR-----TPFPQQPQPQPPQYLQSQ----- 61
Query: 429 QYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQ 472
Q +V+ PQQ QQ QQ Q QQQ + QQQQ+QQ
Sbjct: 62 SDAQQYVQRGYPQQIQQQQQLQ---------QQQQQQQQQQEQQ 96
Score = 32.3 bits (72), Expect = 1.9
Identities = 35/111 (31%), Positives = 47/111 (41%), Gaps = 15/111 (13%)
Query: 382 NAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQ 441
N RF P +GAAG G P Q + P P Q Q LQ + PQ
Sbjct: 7 NRGRFPPG-------IGAAGPGP--DPNFQSRNPNP----PQPQQYLQSRTPFPQQPQPQ 53
Query: 442 QRQQMQQFQHHYRLMQQ-HQQQLRMQQQ-QQQQLHAGVSPVQQPGQVARPP 490
Q +Q + +Q+ + QQ++ QQQ QQQQ QQ + A+ P
Sbjct: 54 PPQYLQSQSDAQQYVQRGYPQQIQQQQQLQQQQQQQQQQQEQQWSRRAQLP 104
>At3g20910 CCAAT-binding factor B chain like protein
Length = 303
Score = 46.6 bits (109), Expect = 1e-04
Identities = 54/245 (22%), Positives = 100/245 (40%), Gaps = 34/245 (13%)
Query: 265 GTTSSESGPLKPWHISARPQNTDS---ANSDISQRIAF-LQTQLAKTPM---PSITSKAE 317
GT SS L PWH +A N +S A S+ IA L++ +K+P ++ +++
Sbjct: 23 GTVSSSINSLNPWHRAAAACNANSSVEAGDKSSKSIALALESNGSKSPSNRDNTVNKESQ 82
Query: 318 LPTIEEAWNTPVVSDWNSKHQPQQKRHPGMAQQGGPKP---NHYVNGVNGGAVPPPPYPG 374
+ T ++ D++ K+ Q+ H G+ Q P P H V + P Y G
Sbjct: 83 VTTSPQS-----AGDYSDKN--QESLHHGITQP-PPHPQLVGHTVGWASSNPYQDPYYAG 134
Query: 375 AVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQ-LLQYYQS 433
+ G + F P G GM H+R P P ++N +Q Q +L+ Q+
Sbjct: 135 VMGAYGHHPLGFVPYG----------GMPHSRMPLPPEMAQEPVFVNAKQYQAILRRRQA 184
Query: 434 HVES-----LNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVAR 488
++ L ++ + + +H + + + R ++ + + + G V +
Sbjct: 185 RAKAELEKKLIKSRKPYLHESRHQHAMRRPRGTGGRFAKKTNTEASKRKAEEKSNGHVTQ 244
Query: 489 PPQQS 493
P S
Sbjct: 245 SPSSS 249
>At1g06070 bZip transcription factor AtbZip69
Length = 423
Score = 45.1 bits (105), Expect = 3e-04
Identities = 30/86 (34%), Positives = 39/86 (44%), Gaps = 5/86 (5%)
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ-QQQQLHAGVS 478
L QQLQ LQ + Q +QQ QQ Q + QQ QL+ QQ+ QQQ+ +G S
Sbjct: 337 LAAQQLQQLQIQSQKQQQQQQQHQQQQQQQQQQFHFQQQQLYQLQQQQRLQQQEQQSGAS 396
Query: 479 ----PVQQPGQVARPPQQSLQQPSVK 500
P+ PGQ + P K
Sbjct: 397 ELRRPMPSPGQKESVTSPDRETPLTK 422
Score = 39.7 bits (91), Expect = 0.012
Identities = 31/89 (34%), Positives = 39/89 (42%), Gaps = 6/89 (6%)
Query: 418 YYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGV 477
+Y N Q + + Q + Q+QQ QQ Q H + QQ QQQ QQQQ QL
Sbjct: 326 FYPNNQSMHTILAAQQLQQLQIQSQKQQQQQ-QQHQQQQQQQQQQFHFQQQQLYQLQQQQ 384
Query: 478 SPVQQPGQ-----VARPPQQSLQQPSVKS 501
QQ Q + RP Q+ SV S
Sbjct: 385 RLQQQEQQSGASELRRPMPSPGQKESVTS 413
Score = 38.1 bits (87), Expect = 0.035
Identities = 26/68 (38%), Positives = 33/68 (48%), Gaps = 1/68 (1%)
Query: 429 QYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM-QQQQQQQLHAGVSPVQQPGQVA 487
Q+Y ++ QQ+QQ Q + QQ QQQ + QQQQQQQ H + Q Q
Sbjct: 325 QFYPNNQSMHTILAAQQLQQLQIQSQKQQQQQQQHQQQQQQQQQQFHFQQQQLYQLQQQQ 384
Query: 488 RPPQQSLQ 495
R QQ Q
Sbjct: 385 RLQQQEQQ 392
Score = 30.0 bits (66), Expect = 9.6
Identities = 23/102 (22%), Positives = 44/102 (42%), Gaps = 5/102 (4%)
Query: 378 GAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVES 437
G+ + ++F PN + + ++ A + + + QQ + QQQ Q Q++ +
Sbjct: 318 GSFGSNQQFYPNNQSMHTILAAQQLQQLQIQSQKQQQQQQQHQQQQQQQQQQFHFQQQQL 377
Query: 438 LNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSP 479
QQ+Q++QQ + Q +LR Q + SP
Sbjct: 378 YQLQQQQRLQQQE-----QQSGASELRRPMPSPGQKESVTSP 414
>At3g11540 spindly (gibberellin signal transduction protein)
Length = 914
Score = 43.9 bits (102), Expect = 6e-04
Identities = 54/239 (22%), Positives = 94/239 (38%), Gaps = 28/239 (11%)
Query: 22 AVRAFQQVLYVDPVFQRANE----VHLRLGLMFKV-NNDPESALKHLKLSYVDASPSTFC 76
A ++Q+ L D ++ A E V LG K+ N E K+ + +D +
Sbjct: 130 AAESYQKALMADASYKPAAECLAIVLTDLGTSLKLAGNTQEGIQKYYEALKIDPHYAP-- 187
Query: 77 KLEIQFHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKT 136
+++ +Y ++ A YE+ E+ P++ A+ Y +G +Y N
Sbjct: 188 ---AYYNLGVVYSEMMQYDNALSCYEKAALER--PMY--AEAYCNMGVIYKN-------R 233
Query: 137 ERMTLAIRCLEKSLE-------ADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTP 189
+ +AI C E+ L A N L LG G Y+ +
Sbjct: 234 GDLEMAITCYERCLAVSPNFEIAKNNMAIALTDLGTKVKLEGDVTQGVAYYKKALYYNWH 293
Query: 190 TADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACY 248
AD ++GV Y + ++ A+ Y A + + + NLG+LY+ L A+ CY
Sbjct: 294 YADAMYNLGVAYGEMLKFDMAIVFYELAFHFNPHCAEACNNLGVLYKDRDNLDKAVECY 352
Score = 43.5 bits (101), Expect = 8e-04
Identities = 28/118 (23%), Positives = 54/118 (45%), Gaps = 7/118 (5%)
Query: 138 RMTLAIRCLEKSLEADRN---SGQTLYL----LGRCYASIGKADHAFDAYRNVVEKTTPT 190
R+ A +K+L AD + + + L + LG G Y ++
Sbjct: 126 RLVEAAESYQKALMADASYKPAAECLAIVLTDLGTSLKLAGNTQEGIQKYYEALKIDPHY 185
Query: 191 ADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACY 248
A + ++GV+Y + QY +AL Y A ++ ++ N+G++Y++ L+ A+ CY
Sbjct: 186 APAYYNLGVVYSEMMQYDNALSCYEKAALERPMYAEAYCNMGVIYKNRGDLEMAITCY 243
Score = 41.2 bits (95), Expect = 0.004
Identities = 48/213 (22%), Positives = 82/213 (37%), Gaps = 37/213 (17%)
Query: 22 AVRAFQQVLYVDPVFQRA-NEVHLRL---GLMFKVNNDPESALKHLKLS------YVDAS 71
A+ +++ L V P F+ A N + + L G K+ D + + K + Y DA
Sbjct: 239 AITCYERCLAVSPNFEIAKNNMAIALTDLGTKVKLEGDVTQGVAYYKKALYYNWHYADA- 297
Query: 72 PSTFCKLEIQFHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLK---ADIYRQLGWMYHN 128
+++ Y K +A YE L H A+ LG +Y +
Sbjct: 298 ---------MYNLGVAYGEMLKFDMAIVFYE-------LAFHFNPHCAEACNNLGVLYKD 341
Query: 129 VEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTT 188
+ + A+ C + +L N Q+L LG Y GK D A +
Sbjct: 342 -------RDNLDKAVECYQMALSIKPNFAQSLNNLGVVYTVQGKMDAAASMIEKAILANP 394
Query: 189 PTADVWCSIGVLYQQQKQYVDALQAYVCAVQLD 221
A+ + ++GVLY+ A+ AY +++D
Sbjct: 395 TYAEAFNNLGVLYRDAGNITMAIDAYEECLKID 427
Score = 37.4 bits (85), Expect = 0.060
Identities = 24/97 (24%), Positives = 44/97 (44%)
Query: 139 MTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIG 198
+T + +K+L + + +Y LG Y + K D A Y A+ ++G
Sbjct: 277 VTQGVAYYKKALYYNWHYADAMYNLGVAYGEMLKFDMAIVFYELAFHFNPHCAEACNNLG 336
Query: 199 VLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILY 235
VLY+ + A++ Y A+ + + S NLG++Y
Sbjct: 337 VLYKDRDNLDKAVECYQMALSIKPNFAQSLNNLGVVY 373
Score = 36.6 bits (83), Expect = 0.10
Identities = 29/148 (19%), Positives = 55/148 (36%), Gaps = 41/148 (27%)
Query: 142 AIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLY 201
A+ E LE D + + G C + K + AFD + + A G+L+
Sbjct: 62 ALALYEAMLEKDSKNVEAHIGKGICLQTQNKGNLAFDCFSEAIRLDPHNACALTHCGILH 121
Query: 202 QQQKQYVDA-----------------------------------------LQAYVCAVQL 220
+++ + V+A +Q Y A+++
Sbjct: 122 KEEGRLVEAAESYQKALMADASYKPAAECLAIVLTDLGTSLKLAGNTQEGIQKYYEALKI 181
Query: 221 DKCHSASWINLGILYESCSQLKDALACY 248
D ++ ++ NLG++Y Q +AL+CY
Sbjct: 182 DPHYAPAYYNLGVVYSEMMQYDNALSCY 209
Score = 33.9 bits (76), Expect = 0.66
Identities = 28/133 (21%), Positives = 54/133 (40%), Gaps = 1/133 (0%)
Query: 125 MYHNVEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVV 184
MY+ A+G + +AI E + + + + LG Y D A + Y+ +
Sbjct: 298 MYNLGVAYGEML-KFDMAIVFYELAFHFNPHCAEACNNLGVLYKDRDNLDKAVECYQMAL 356
Query: 185 EKTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDA 244
A ++GV+Y Q + A A+ + ++ ++ NLG+LY + A
Sbjct: 357 SIKPNFAQSLNNLGVVYTVQGKMDAAASMIEKAILANPTYAEAFNNLGVLYRDAGNITMA 416
Query: 245 LACYIHSVRASKD 257
+ Y ++ D
Sbjct: 417 IDAYEECLKIDPD 429
Score = 30.4 bits (67), Expect = 7.3
Identities = 24/92 (26%), Positives = 46/92 (49%), Gaps = 4/92 (4%)
Query: 167 YASIGKADHAF-DA---YRNVVEKTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDK 222
YA+I +A + F DA Y ++EK + + G+ Q Q + A + A++LD
Sbjct: 49 YANILRARNKFADALALYEAMLEKDSKNVEAHIGKGICLQTQNKGNLAFDCFSEAIRLDP 108
Query: 223 CHSASWINLGILYESCSQLKDALACYIHSVRA 254
++ + + GIL++ +L +A Y ++ A
Sbjct: 109 HNACALTHCGILHKEEGRLVEAAESYQKALMA 140
>At1g49600 DNA binding protein ACBF, putative
Length = 445
Score = 43.5 bits (101), Expect = 8e-04
Identities = 32/98 (32%), Positives = 44/98 (44%), Gaps = 13/98 (13%)
Query: 389 NGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVE-----SLNPQQR 443
NG D+ ++ G PP Q PPP QQQ Q Q + + ++ ++ Q+
Sbjct: 7 NGSTDS-VLPPTSAGTTPPPPLQQSTPPPQQQQQQQWQQQQQWMAAMQQYPAAAMAMMQQ 65
Query: 444 QQMQQFQH------HYRLMQQHQQ-QLRMQQQQQQQLH 474
QQM + H + QQH Q Q QQQQQQ H
Sbjct: 66 QQMMMYPHPQYAPYNQAAYQQHPQFQYAAYQQQQQQHH 103
Score = 31.2 bits (69), Expect = 4.3
Identities = 28/92 (30%), Positives = 36/92 (38%), Gaps = 21/92 (22%)
Query: 407 PPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQ----Q 462
PP + PPP LQ +S P Q+QQ QQ+Q + M QQ
Sbjct: 15 PPTSAGTTPPP------PLQ---------QSTPPPQQQQQQQWQQQQQWMAAMQQYPAAA 59
Query: 463 LRMQQQQQQQL--HAGVSPVQQPGQVARPPQQ 492
+ M QQQQ + H +P Q P Q
Sbjct: 60 MAMMQQQQMMMYPHPQYAPYNQAAYQQHPQFQ 91
>At1g17440 unknown protein
Length = 683
Score = 43.5 bits (101), Expect = 8e-04
Identities = 36/98 (36%), Positives = 45/98 (45%), Gaps = 6/98 (6%)
Query: 406 RPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQ-----RQQMQQFQHHYRLMQQHQ 460
+P Q Q QQQL Q +QS + SLN QQ +QQ QQ Q M Q
Sbjct: 388 QPSQRQALLLQQQQQQQQQLSSPQLHQSSM-SLNQQQISQIIQQQQQQSQLGQSQMNQSH 446
Query: 461 QQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPS 498
Q ++QQ QQQ +QQ Q + Q + QQPS
Sbjct: 447 SQQQLQQMQQQLQQQPQQQMQQQQQQQQQMQINQQQPS 484
Score = 41.2 bits (95), Expect = 0.004
Identities = 52/191 (27%), Positives = 79/191 (41%), Gaps = 45/191 (23%)
Query: 305 AKTPMPSITSKAELPTIEEAWNTPVVSDWNSKHQPQQK--RHPGMAQQGGPKPNHYVNGV 362
+ TP+PS +S+ + + P+ + NS P G+ Q +PN ++
Sbjct: 27 SSTPLPSSSSQQQ-----QLMTAPISNSVNSAASPAMTVTTTEGIVIQNNSQPN--ISSP 79
Query: 363 NGGAVPPPPYPGAVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQ--PPPYYL 420
N + PP +P P + P+ +D Q Q QQ L
Sbjct: 80 NPTS-SNPPIGAQIPSPSPLSH---PSSSLDQ--------------QTQTQQLVQQTQQL 121
Query: 421 NQQQLQLLQYYQSH-VESLNPQQRQQMQQFQH---------HYRLMQQHQQ-----QLRM 465
QQQ Q++Q S + L+PQQ+Q +QQ QH Y++ Q Q+ +L
Sbjct: 122 PQQQQQIMQQISSSPIPQLSPQQQQILQQ-QHMTSQQIPMSSYQIAQSLQRSPSLSRLSQ 180
Query: 466 QQQQQQQLHAG 476
QQQQQQ H G
Sbjct: 181 IQQQQQQQHQG 191
Score = 40.8 bits (94), Expect = 0.005
Identities = 31/96 (32%), Positives = 38/96 (39%), Gaps = 2/96 (2%)
Query: 411 QPQQPPPYYLNQQQLQLLQYY--QSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
QP Q L QQQ Q Q Q H S++ Q+Q Q Q + Q Q Q+
Sbjct: 388 QPSQRQALLLQQQQQQQQQLSSPQLHQSSMSLNQQQISQIIQQQQQQSQLGQSQMNQSHS 447
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPP 504
QQQ QQP Q + QQ QQ + P
Sbjct: 448 QQQLQQMQQQLQQQPQQQMQQQQQQQQQMQINQQQP 483
Score = 35.8 bits (81), Expect = 0.17
Identities = 32/89 (35%), Positives = 37/89 (40%), Gaps = 9/89 (10%)
Query: 422 QQQLQLLQYYQSHV-ESLNPQQRQQMQQ-FQHHYRLMQQHQQQLRMQQQQQQQ------- 472
QQQ Q Q QS + +S + QQ QQMQQ Q + Q QQQ + Q Q QQ
Sbjct: 429 QQQQQQSQLGQSQMNQSHSQQQLQQMQQQLQQQPQQQMQQQQQQQQQMQINQQQPSPRML 488
Query: 473 LHAGVSPVQQPGQVARPPQQSLQQPSVKS 501
HAG V G Q P S
Sbjct: 489 SHAGQKSVSLTGSQPEATQSGTTTPGGSS 517
Score = 31.6 bits (70), Expect = 3.3
Identities = 27/90 (30%), Positives = 43/90 (47%), Gaps = 13/90 (14%)
Query: 419 YLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQ-----HHYRLMQQHQQQLRMQQQQQQ-- 471
++N Q L Q Q+ + + Q+Q ++Q + +RL +Q L +QQQQQQ
Sbjct: 346 FMNPQLSGLAQNGQAGMMQNSLSQQQWLKQMSGITSPNSFRLQPSQRQALLLQQQQQQQQ 405
Query: 472 -----QLHAGVSPVQQPGQVARPPQQSLQQ 496
QLH + Q Q+++ QQ QQ
Sbjct: 406 QLSSPQLHQSSMSLNQQ-QISQIIQQQQQQ 434
Score = 30.8 bits (68), Expect = 5.6
Identities = 26/69 (37%), Positives = 31/69 (44%), Gaps = 12/69 (17%)
Query: 433 SHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQ 492
SH S QQ Q Q L+QQ QQ + QQQ QQ+ + P P Q QQ
Sbjct: 99 SHPSSSLDQQTQTQQ-------LVQQTQQLPQQQQQIMQQISSSPIPQLSPQQ-----QQ 146
Query: 493 SLQQPSVKS 501
LQQ + S
Sbjct: 147 ILQQQHMTS 155
>At3g15000 unknown protein
Length = 395
Score = 43.1 bits (100), Expect = 0.001
Identities = 27/81 (33%), Positives = 31/81 (37%), Gaps = 6/81 (7%)
Query: 345 PGMAQQGGPKPNHYVNGVN------GGAVPPPPYPGAVPGAGPNAKRFKPNGEVDNGMVG 398
P GGP P ++ G GG+ PPPP+ G G P P G
Sbjct: 243 PQRPPMGGPPPPPHIGGSAPPPPHMGGSAPPPPHMGQNYGPPPPNNMGGPRHPPPYGAPP 302
Query: 399 AAGMGHARPPQAQPQQPPPYY 419
MG RPPQ PPP Y
Sbjct: 303 QNNMGGPRPPQNYGGTPPPNY 323
Score = 35.4 bits (80), Expect = 0.23
Identities = 35/138 (25%), Positives = 49/138 (35%), Gaps = 33/138 (23%)
Query: 319 PTIEEAWNTPVVSDWNSKHQPQQKRHPGMAQQGGPKPNHYVNGVNGGAVPPPPYPGAVPG 378
P + + + P ++ P P GGP+P N G PPP Y GA P
Sbjct: 275 PHMGQNYGPPPPNNMGGPRHPPPYGAPPQNNMGGPRPPQ-----NYGGTPPPNYGGAPP- 328
Query: 379 AGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESL 438
A MG A PP PP Y + QY + ++
Sbjct: 329 --------------------ANNMGGAPPPNYGGGPPPQY----GAVPPPQYGGAPPQNN 364
Query: 439 NPQQR---QQMQQFQHHY 453
N QQ+ Q Q+Q++Y
Sbjct: 365 NYQQQGSGMQQPQYQNNY 382
>At1g15780 unknown protein
Length = 1335
Score = 42.7 bits (99), Expect = 0.001
Identities = 28/56 (50%), Positives = 31/56 (55%), Gaps = 3/56 (5%)
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHA 475
+NQ QL H++ QQ Q QQFQ R MQQ Q Q R QQQQQQQL A
Sbjct: 815 VNQPQLSSSLLQHQHLKQQQDQQMQLKQQFQQ--RQMQQQQLQAR-QQQQQQQLQA 867
Score = 42.7 bits (99), Expect = 0.001
Identities = 33/96 (34%), Positives = 43/96 (44%), Gaps = 14/96 (14%)
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQMQQF-------QHHYRLMQQHQQQLRMQQQQQQQ 472
L QQ QL++ Q S QQ+Q M Q QH RL+ Q + +QQQQ QQ
Sbjct: 351 LQQQPTQLMR--QQAANSSGIQQKQMMGQHVVGDMQQQHQQRLLNQQNNVMNIQQQQSQQ 408
Query: 473 LHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVH 508
P+QQP Q + + QQ + QA H
Sbjct: 409 -----QPLQQPQQQQKQQPPAQQQLMSQQNSLQATH 439
Score = 37.7 bits (86), Expect = 0.046
Identities = 33/116 (28%), Positives = 47/116 (40%), Gaps = 14/116 (12%)
Query: 406 RPPQAQPQQPP--------PYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQ 457
+P Q Q P P +N LQ L Q L+ QQ Q QH +++
Sbjct: 266 QPNQLHSSQQPGVPTSATQPSTVNSAPLQGLHTNQQSSPQLSSQQTTQSMLRQHQSSMLR 325
Query: 458 QH---QQQLRMQQQQQQQLHAGVSPV-QQPGQVARPPQQSLQQPSVKSCPPQAVHI 509
QH QQ + QQQ +SP+ QQP Q+ R QQ+ ++ H+
Sbjct: 326 QHPQSQQASGIHQQQSSLPQQSISPLQQQPTQLMR--QQAANSSGIQQKQMMGQHV 379
Score = 37.4 bits (85), Expect = 0.060
Identities = 32/107 (29%), Positives = 42/107 (38%), Gaps = 20/107 (18%)
Query: 402 MGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQ 461
+G +Q QQ P Y Q Q QLL +Q Q + L+ H Q
Sbjct: 211 LGRPHAMSSQQQQQPYLYQQQLQQQLL------------KQNFQSGNVPNPNSLLPSHIQ 258
Query: 462 QLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVH 508
Q + Q QLH+ QQPG P + Q +V S P Q +H
Sbjct: 259 QQQQNVLQPNQLHSS----QQPG----VPTSATQPSTVNSAPLQGLH 297
Score = 36.6 bits (83), Expect = 0.10
Identities = 30/100 (30%), Positives = 40/100 (40%), Gaps = 2/100 (2%)
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQ--MQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGV 477
+ QQ Q L Q++V ++ QQ QQ +QQ Q + QQQL QQ Q H
Sbjct: 383 MQQQHQQRLLNQQNNVMNIQQQQSQQQPLQQPQQQQKQQPPAQQQLMSQQNSLQATHQNP 442
Query: 478 SPVQQPGQVARPPQQSLQQPSVKSCPPQAVHIPFSFLSTP 517
Q + PQQ + V + Q LS P
Sbjct: 443 LGTQSNVAGLQQPQQQMLNSQVGNSSLQNNQHSVHMLSQP 482
Score = 35.0 bits (79), Expect = 0.30
Identities = 32/114 (28%), Positives = 47/114 (41%), Gaps = 11/114 (9%)
Query: 387 KPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLN-PQQRQQ 445
+P ++ N VG + + + + QP Q Y +S N P Q+Q
Sbjct: 454 QPQQQMLNSQVGNSSLQNNQHSVHMLSQPTVGLQRTHQAGHGLYSSQGQQSQNQPSQQQM 513
Query: 446 MQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVA---RPPQQSLQQ 496
M Q Q H++ + QQ +QQ QQ+L A GQV PPQ + Q
Sbjct: 514 MPQLQSHHQQLGLQQQPNLLQQDVQQRLQAS-------GQVTGSLLPPQNVVDQ 560
Score = 33.5 bits (75), Expect = 0.86
Identities = 57/249 (22%), Positives = 93/249 (36%), Gaps = 49/249 (19%)
Query: 291 SDISQRIAFLQTQLAKTPMPSITSKAEL-PTIEE--AWNTPVVSDWNSKHQPQQKRHPGM 347
SD +++ +T L + SK+ + P +++ A+ + + + H+P++ G
Sbjct: 634 SDQLEKLRQFKTMLERMIQFLSVSKSNIMPALKDKVAYYEKQIIGFLNMHRPRKPVQQGQ 693
Query: 348 AQQGGPKPNHYVNGV------NGGAVPPPPYPGAVPGAGPNAKRFKPNGEVDNGMVGAAG 401
Q +P + P ++ GAGP A++ N + G
Sbjct: 694 LPQSQMQPMQQPQSQTVQDQSHDNQTNPQMQSMSMQGAGPRAQQSSMTNMQSNVLSSRPG 753
Query: 402 MGHARPPQAQPQQPPPYYL---------NQQQLQLLQYYQSHVESLN------------- 439
+ + P Q P P L N QQ+ + Q+ + +N
Sbjct: 754 V--SAPQQNIPSSIPASSLESGQGNTLNNGQQVAMGSMQQNTSQLVNNSSASAQSGLSTL 811
Query: 440 ------PQQRQQMQQFQHHYRLMQQHQQQLRMQQQ------QQQQLHAGVSPVQQPGQVA 487
PQ + Q QH L QQ QQ++++QQ QQQQL A QQ Q A
Sbjct: 812 QSNVNQPQLSSSLLQHQH---LKQQQDQQMQLKQQFQQRQMQQQQLQARQQQQQQQLQ-A 867
Query: 488 RPPQQSLQQ 496
R LQQ
Sbjct: 868 RQQAAQLQQ 876
Score = 33.1 bits (74), Expect = 1.1
Identities = 34/109 (31%), Positives = 43/109 (39%), Gaps = 14/109 (12%)
Query: 406 RPPQAQPQQPPPYYLNQQQLQLLQY-------YQSHVESLNPQQRQQMQQFQHHYRLMQQ 458
+P Q Q QQPP Q LQ QS+V L Q +QQM Q +Q
Sbjct: 413 QPQQQQKQQPPAQQQLMSQQNSLQATHQNPLGTQSNVAGLQ-QPQQQMLNSQVGNSSLQN 471
Query: 459 HQQQLRMQQQQQQQLHAGVSPVQQPGQ--VARPPQQSLQQPSVKSCPPQ 505
+Q + M Q G+ Q G + QQS QPS + PQ
Sbjct: 472 NQHSVHMLSQPT----VGLQRTHQAGHGLYSSQGQQSQNQPSQQQMMPQ 516
Score = 33.1 bits (74), Expect = 1.1
Identities = 26/68 (38%), Positives = 32/68 (46%), Gaps = 8/68 (11%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQ-QLRMQ 466
PQ +L QQQ Q +Q Q Q++QMQQ Q R QQ QQ Q R Q
Sbjct: 818 PQLSSSLLQHQHLKQQQDQQMQLKQQF-------QQRQMQQQQLQARQQQQQQQLQARQQ 870
Query: 467 QQQQQQLH 474
Q QQ++
Sbjct: 871 AAQLQQMN 878
>At5g56290 peroxisomal targeting signal type 1 receptor
Length = 728
Score = 42.4 bits (98), Expect = 0.002
Identities = 21/91 (23%), Positives = 42/91 (46%)
Query: 162 LLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLD 221
+LG Y + D A +++ ++ +W +G Q DA+ AY A+ L
Sbjct: 595 VLGVLYNLSREFDRAITSFQTALQLKPNDYSLWNKLGATQANSVQSADAISAYQQALDLK 654
Query: 222 KCHSASWINLGILYESCSQLKDALACYIHSV 252
+ +W N+GI Y + K+++ Y+ ++
Sbjct: 655 PNYVRAWANMGISYANQGMYKESIPYYVRAL 685
Score = 38.5 bits (88), Expect = 0.027
Identities = 32/133 (24%), Positives = 58/133 (43%), Gaps = 9/133 (6%)
Query: 116 ADIYRQLGWMYHNVEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADH 175
AD++ LG +Y+ F AI + +L+ N LG A+ ++
Sbjct: 590 ADVHIVLGVLYNLSREFDR-------AITSFQTALQLKPNDYSLWNKLGATQANSVQSAD 642
Query: 176 AFDAYRNVVEKTTPTADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILY 235
A AY+ ++ W ++G+ Y Q Y +++ YV A+ ++ +W L L
Sbjct: 643 AISAYQQALDLKPNYVRAWANMGISYANQGMYKESIPYYVRALAMNPKADNAWQYLR-LS 701
Query: 236 ESCSQLKDAL-AC 247
SC+ +D + AC
Sbjct: 702 LSCASRQDMIEAC 714
Score = 36.6 bits (83), Expect = 0.10
Identities = 26/105 (24%), Positives = 48/105 (44%), Gaps = 4/105 (3%)
Query: 191 ADVWCSIGVLYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIH 250
ADV +GVLY +++ A+ ++ A+QL + W LG + Q DA++ Y
Sbjct: 590 ADVHIVLGVLYNLSREFDRAITSFQTALQLKPNDYSLWNKLGATQANSVQSADAISAYQQ 649
Query: 251 SVRASKDKA-AGVGSGTTSSESGPLK---PWHISARPQNTDSANS 291
++ + A G + + G K P+++ A N + N+
Sbjct: 650 ALDLKPNYVRAWANMGISYANQGMYKESIPYYVRALAMNPKADNA 694
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,717,008
Number of Sequences: 26719
Number of extensions: 1450785
Number of successful extensions: 11288
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 81
Number of HSP's successfully gapped in prelim test: 129
Number of HSP's that attempted gapping in prelim test: 8118
Number of HSP's gapped (non-prelim): 1488
length of query: 1366
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1255
effective length of database: 8,352,787
effective search space: 10482747685
effective search space used: 10482747685
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC139744.2


