

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.6 - phase: 0
(272 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
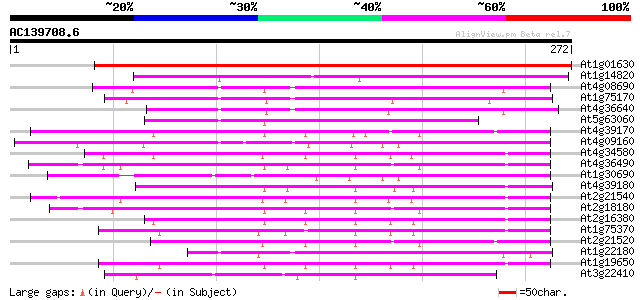
Score E
Sequences producing significant alignments: (bits) Value
At1g01630 polyphosphoinositide binding protein, putative 322 2e-88
At1g14820 SEC14, phosphatidylinositol/phosphatidylcholine transf... 159 2e-39
At4g08690 putative phosphoglyceride transfer protein 115 2e-26
At1g75170 unknown protein 108 4e-24
At4g36640 unknown protein 101 5e-22
At5g63060 unknown protein 99 2e-21
At4g39170 unknown protein 95 5e-20
At4g09160 putative protein 94 6e-20
At4g34580 putative protein 93 1e-19
At4g36490 hypothetical protein 93 2e-19
At1g30690 putative phosphatidyl-inositol-transfer protein 92 2e-19
At4g39180 SEC14 - like protein 91 7e-19
At2g21540 putative phosphatidylinositol/phophatidylcholine trans... 89 2e-18
At2g18180 putative phosphatidylinositol/phophatidylcholine trans... 89 3e-18
At2g16380 putative phosphatidylinositol/phosphatidylcholine tran... 88 4e-18
At1g75370 sec14 cytosolic factor, putative 87 1e-17
At2g21520 putative phosphatidylinositol/phosphatidylcholine tran... 86 2e-17
At1g22180 unknown protein 84 1e-16
At1g19650 sec14 cytosolic factor- like protein 83 2e-16
At3g22410 unknown protein 82 3e-16
>At1g01630 polyphosphoinositide binding protein, putative
Length = 255
Score = 322 bits (824), Expect = 2e-88
Identities = 151/231 (65%), Positives = 188/231 (81%)
Query: 42 EAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPS 101
E E +K+ +MR L + +DP +KEVDDLMIRRFLRARDLD++KAS MFL Y+ W++S +P
Sbjct: 25 EIERSKVGIMRALCDRQDPETKEVDDLMIRRFLRARDLDIEKASTMFLNYLTWKRSMLPK 84
Query: 102 GSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLIS 161
G + +EIA+DL+ K+ +QG DK GRPI VA +H +K D FKR+VV+ LEK+ +
Sbjct: 85 GHIPEAEIANDLSHNKMCMQGHDKMGRPIAVAIGNRHNPSKGNPDEFKRFVVYTLEKICA 144
Query: 162 RMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWK 221
RMP G+EKFV+I D++GWGY+N DIRGYL AL+ LQD YPERLGKL+IVHAPY+FM WK
Sbjct: 145 RMPRGQEKFVAIGDLQGWGYSNCDIRGYLAALSTLQDCYPERLGKLYIVHAPYIFMTAWK 204
Query: 222 IIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQDS 272
+IYPFID NTKKKIVFVENKKL TLLE+IDESQLP+IYGGKLPLVPIQ++
Sbjct: 205 VIYPFIDANTKKKIVFVENKKLTPTLLEDIDESQLPDIYGGKLPLVPIQET 255
>At1g14820 SEC14, phosphatidylinositol/phosphatidylcholine transfer
protein, putative
Length = 239
Score = 159 bits (401), Expect = 2e-39
Identities = 87/216 (40%), Positives = 127/216 (58%), Gaps = 6/216 (2%)
Query: 61 SSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVP-SGSVSPSEIADDLAQEKIY 119
S K D + RFL AR +D KA+ MF+ + KWR S VP +G + SE+ D+L K+
Sbjct: 9 SEKGYDKPTLMRFLVARSMDPVKAAKMFVDWQKWRASMVPPTGFIPESEVQDELEFRKVC 68
Query: 120 VQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEE----KFVSIAD 175
+QG K G P+++ +KHF +K+ + FK++VV+AL+K I+ G+E K V++ D
Sbjct: 69 LQGPTKSGHPLVLVITSKHFASKDPAN-FKKFVVYALDKTIASGNNGKEVGGEKLVAVID 127
Query: 176 IKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKI 235
+ Y N D RG + LQ YYPERL K +I+H P F+ VWK + F++ T++KI
Sbjct: 128 LANITYKNLDARGLITGFQFLQSYYPERLAKCYILHMPGFFVTVWKFVCRFLEKATQEKI 187
Query: 236 VFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQD 271
V V + + + EEI LPE YGG+ L IQD
Sbjct: 188 VIVTDGEEQRKFEEEIGADALPEEYGGRAKLTAIQD 223
>At4g08690 putative phosphoglyceride transfer protein
Length = 301
Score = 115 bits (288), Expect = 2e-26
Identities = 70/226 (30%), Positives = 115/226 (49%), Gaps = 7/226 (3%)
Query: 41 TEAELTKINLMRTLVESR-DPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFV 99
TE E KI +R L+ + S D + R+LRAR+ V KA+ M + +KWR +
Sbjct: 16 TEEEQAKIEEVRKLLGPLPEKLSSFCSDDAVLRYLRARNWHVKKATKMLKETLKWRVQYK 75
Query: 100 PSGSVSPSEIADDLAQEKIYVQG-LDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEK 158
P + E+A + KIY +DK GRP+++ + +N + RY+V+ +E
Sbjct: 76 PE-EICWEEVAGEAETGKIYRSSCVDKLGRPVLIMRPS--VENSKSVKGQIRYLVYCMEN 132
Query: 159 LISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMK 218
+ +PPGEE+ V + D G+ AN +R +LQ++YPERL + + P F
Sbjct: 133 AVQNLPPGEEQMVWMIDFHGYSLANVSLRTTKETAHVLQEHYPERLAFAVLYNPPKFFEP 192
Query: 219 VWKIIYPFIDDNTKKKIVFV--ENKKLKATLLEEIDESQLPEIYGG 262
WK+ PF++ T+ K+ FV ++ K + E D ++ +GG
Sbjct: 193 FWKVARPFLEPKTRNKVKFVYSDDPNTKVIMEENFDMEKMELAFGG 238
>At1g75170 unknown protein
Length = 296
Score = 108 bits (269), Expect = 4e-24
Identities = 68/222 (30%), Positives = 115/222 (51%), Gaps = 8/222 (3%)
Query: 47 KINLMRTLV-ESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVS 105
K+ ++TL+ + +S D ++R+L AR+ +V KA M + +KWR SF P +
Sbjct: 23 KMKELKTLIGQLSGRNSLYCSDACLKRYLEARNWNVGKAKKMLEETLKWRSSFKPE-EIR 81
Query: 106 PSEIADDLAQEKIYVQGL-DKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMP 164
+E++ + K+Y G D+ GR +++ QN L+ +++V+ +E I +P
Sbjct: 82 WNEVSGEGETGKVYKAGFHDRHGRTVLILRPG--LQNTKSLENQMKHLVYLIENAILNLP 139
Query: 165 PGEEKFVSIADIKGWGYANS-DIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKII 223
+E+ + D GW + S I+ + ILQ++YPERL F+ + P +F WKI+
Sbjct: 140 EDQEQMSWLIDFTGWSMSTSVPIKSARETINILQNHYPERLAVAFLYNPPRLFEAFWKIV 199
Query: 224 YPFIDDNT--KKKIVFVENKKLKATLLEEIDESQLPEIYGGK 263
FID T K K V+ +N + + DE LP +GGK
Sbjct: 200 KYFIDAKTFVKVKFVYPKNSESVELMSTFFDEENLPTEFGGK 241
>At4g36640 unknown protein
Length = 294
Score = 101 bits (251), Expect = 5e-22
Identities = 63/204 (30%), Positives = 107/204 (51%), Gaps = 7/204 (3%)
Query: 67 DLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGL-DK 125
D +RRFL AR+ DV+KA M + +KWR ++ P + +++A + K D+
Sbjct: 41 DASLRRFLDARNWDVEKAKKMIQETLKWRSTYKPQ-EIRWNQVAHEGETGKASRASFHDR 99
Query: 126 KGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYA-NS 184
+GR +++ A QN + R++V+ LE I +P G+++ + D GW A N
Sbjct: 100 QGRVVLIMRPA--MQNSTSQEGNIRHLVYLLENAIINLPKGQKQMSWLIDFTGWSMAVNP 157
Query: 185 DIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFV--ENKK 242
++ + ILQ+YYPERLG F+ + P +F V++ F+D T +K+ FV ++K
Sbjct: 158 PMKTTREIIHILQNYYPERLGIAFLYNPPRLFQAVYRAAKYFLDPRTAEKVKFVYPKDKA 217
Query: 243 LKATLLEEIDESQLPEIYGGKLPL 266
+ D LP+ +GG+ L
Sbjct: 218 SDELMTTHFDVENLPKEFGGEATL 241
>At5g63060 unknown protein
Length = 263
Score = 99.4 bits (246), Expect = 2e-21
Identities = 55/163 (33%), Positives = 84/163 (50%), Gaps = 2/163 (1%)
Query: 66 DDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG-LD 124
D+ MI FL+ R VD+A K +KWR F +S I K YV G LD
Sbjct: 71 DEDMILWFLKDRRFSVDEAIGKLTKAIKWRHEFKVD-ELSEDSIKAATDTGKAYVHGFLD 129
Query: 125 KKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYANS 184
KGRP+++ AKH ++ VF LEK +S++P G+ K + I D++G+G N+
Sbjct: 130 VKGRPVVIVAPAKHIPGLLDPIEDEKLCVFLLEKALSKLPAGQHKILGIFDLRGFGSQNA 189
Query: 185 DIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFI 227
D++ + YYP RL ++ V AP++F +W+ P +
Sbjct: 190 DLKFLTFLFDVFYYYYPSRLDEVLFVDAPFIFQPIWQFTKPLV 232
>At4g39170 unknown protein
Length = 614
Score = 94.7 bits (234), Expect = 5e-20
Identities = 81/272 (29%), Positives = 125/272 (45%), Gaps = 22/272 (8%)
Query: 11 NSALDSKHELAKEETKGKILVQDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDL-M 69
N++ KH L K+ K + V + EL ++ R + + + DD M
Sbjct: 49 NASTKFKHSLKKKRRKSDVRVSSVSIEDVRDVEELQAVDEFRQALVMEELLPHKHDDYHM 108
Query: 70 IRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG---LDKK 126
+ RFL+AR D++KA M+ ++WRK F + + + K Y G +DK+
Sbjct: 109 MLRFLKARKFDIEKAKHMWADMIQWRKEFGTDTIIQDFQFEEIDEVLKYYPHGYHSVDKE 168
Query: 127 GRPIIVAFAAKHFQNK----NGLDAFKRYVVFALEKLISRMPP----GEEKFV----SIA 174
GRP+ + K NK LD + RY V E+ P +K++ +I
Sbjct: 169 GRPVYIERLGKVDPNKLMQVTTLDRYIRYHVKEFERSFMLKFPACTIAAKKYIDSSTTIL 228
Query: 175 DIKGWGYANSDIRGYLGALTILQ----DYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDN 230
D++G G N + +T LQ D YPE L ++FI++A F +W + F+D
Sbjct: 229 DVQGVGLKNF-TKSARELITRLQKIDGDNYPETLHQMFIINAGPGFRLLWSTVKSFLDPK 287
Query: 231 TKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
T KI V K ++ LLE ID S+LPE GG
Sbjct: 288 TTSKI-HVLGCKYQSKLLEIIDSSELPEFLGG 318
>At4g09160 putative protein
Length = 723
Score = 94.4 bits (233), Expect = 6e-20
Identities = 80/294 (27%), Positives = 136/294 (46%), Gaps = 37/294 (12%)
Query: 3 TVNPEPGRNSALDSKHELAKEETKGKILV--------QDDGALNDSTEAELTKINLMRTL 54
T + R+ A E A+ E K KI + ++ ++D +E EL + +R L
Sbjct: 256 TTSTVASRSLAEMMNREEAEVEEKQKIQIPRSLGSFKEETNKISDLSETELNALQELRHL 315
Query: 55 VESRDPSSK-----------EVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGS 103
++ SSK + D+++ +FLRARD +A +M K ++WR F
Sbjct: 316 LQVSQDSSKTSIWGVPLLKDDRTDVVLLKFLRARDFKPQEAYSMLNKTLQWRIDFNIE-E 374
Query: 104 VSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKN----------GLDAFKRYVV 153
+ + DDL + +++QG DK+ P+ + FQNK+ + F R+ +
Sbjct: 375 LLDENLGDDL-DKVVFMQGQDKENHPVCYNVYGE-FQNKDLYQKTFSDEEKRERFLRWRI 432
Query: 154 FALEKLISRMP---PGEEKFVSIADIKGW-GYANSDIR-GYLGALTILQDYYPERLGKLF 208
LEK I + G + D+K G +++R AL +LQD YPE + K
Sbjct: 433 QFLEKSIRNLDFVAGGVSTICQVNDLKNSPGPGKTELRLATKQALHLLQDNYPEFVSKQI 492
Query: 209 IVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
++ P+ ++ ++II PF+ +K K+VF + TLL+ I +P YGG
Sbjct: 493 FINVPWWYLAFYRIISPFMSQRSKSKLVFAGPSRSAETLLKYISPEHVPVQYGG 546
>At4g34580 putative protein
Length = 560
Score = 93.2 bits (230), Expect = 1e-19
Identities = 76/246 (30%), Positives = 118/246 (47%), Gaps = 21/246 (8%)
Query: 37 LNDSTEAE-LTKINLMRTLVESRDPSSKEVDDL-MIRRFLRARDLDVDKASAMFLKYMKW 94
+ D +AE L ++ R + + ++DDL M+ RFLRAR D++KA M+ ++W
Sbjct: 54 IEDDIDAEDLQALDAFRQALILDELLPSKLDDLHMMLRFLRARKFDIEKAKQMWSDMIQW 113
Query: 95 RKSFVPSGSVSPSEIADDLAQEKIYVQ---GLDKKGRPIIVAFAAKHFQNK----NGLDA 147
RK F + + + K Y Q G+DK+GRP+ + + NK +D
Sbjct: 114 RKDFGADTIIEDFDFEEIDEVMKHYPQGYHGVDKEGRPVYIERLGQIDANKLLQVTTMDR 173
Query: 148 FKRYVVFALEKLISRMPPG--------EEKFVSIADIKGWGYAN--SDIRGYLGAL-TIL 196
+ +Y V EK P ++ +I D++G G N R L L I
Sbjct: 174 YVKYHVKEFEKTFKVKFPSCSVAANKHIDQSTTILDVQGVGLKNFSKSARELLQRLCKID 233
Query: 197 QDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQL 256
+ YPE L ++FI++A F +W + F+D T KI + N K + LLE ID S+L
Sbjct: 234 NENYPETLNRMFIINAGSGFRLLWSTVKSFLDPKTTAKIHVLGN-KYHSKLLEVIDASEL 292
Query: 257 PEIYGG 262
PE +GG
Sbjct: 293 PEFFGG 298
>At4g36490 hypothetical protein
Length = 543
Score = 92.8 bits (229), Expect = 2e-19
Identities = 76/274 (27%), Positives = 130/274 (46%), Gaps = 25/274 (9%)
Query: 10 RNSALDSKHELAKEETKGKILVQDDGALNDSTEAE-LTKINLMR-TLVESRDPSSKEVDD 67
R+S+ + ++ + K K++ + + D +AE L ++ R +L+ K D
Sbjct: 19 RSSSKNLRYSMTKRRRSSKVMSVE--IIEDVHDAEELKAVDAFRQSLILDELLPEKHDDY 76
Query: 68 LMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG---LD 124
M+ RFL+AR D++K M+ + ++WRK F + + + K Y QG +D
Sbjct: 77 HMMLRFLKARKFDLEKTKQMWTEMLRWRKEFGADTVMEEFDFKEIDEVLKYYPQGHHGVD 136
Query: 125 KKGRPIIVA----FAAKHFQNKNGLDAFKRYVVFALEKLISRMPPG--------EEKFVS 172
K+GRP+ + + +D + Y V E+ + P ++ +
Sbjct: 137 KEGRPVYIERLGLVDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACSIAAKKHIDQSTT 196
Query: 173 IADIKGWGYANSDIRGYLGALTILQ----DYYPERLGKLFIVHAPYMFMKVWKIIYPFID 228
I D++G G N + + +T LQ D YPE L ++FI++A F +W + F+D
Sbjct: 197 ILDVQGVGLKNFN-KAARDLITRLQKVDGDNYPETLNRMFIINAGSGFRMLWNTVKSFLD 255
Query: 229 DNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
T KI + N K ++ LLE IDES+LPE GG
Sbjct: 256 PKTTAKIHVLGN-KYQSKLLEIIDESELPEFLGG 288
>At1g30690 putative phosphatidyl-inositol-transfer protein
Length = 540
Score = 92.4 bits (228), Expect = 2e-19
Identities = 70/253 (27%), Positives = 120/253 (46%), Gaps = 18/253 (7%)
Query: 19 ELAKEETKGKILVQDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARD 78
++ EE K + + +D + + EL + L+ + E D+++ +FLRARD
Sbjct: 179 DVVTEEVKAETIEVEDEDESVDKDIELWGVPLLPS-------KGAESTDVILLKFLRARD 231
Query: 79 LDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKH 138
V++A M K +KWRK S+ E +DLA Y+ G+D++ P+ ++
Sbjct: 232 FKVNEAFEMLKKTLKWRKQN-KIDSILGEEFGEDLATAA-YMNGVDRESHPVCYNVHSEE 289
Query: 139 FQNKNGLDA----FKRYVVFALEKLISRM---PPGEEKFVSIADIKGW-GYANSDIR-GY 189
G + F R+ +EK I ++ P G + I D+K G + ++I G
Sbjct: 290 LYQTIGSEKNREKFLRWRFQLMEKGIQKLNLKPGGVTSLLQIHDLKNAPGVSRTEIWVGI 349
Query: 190 LGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLE 249
+ LQD YPE + + ++ P+ F + ++ PF+ TK K V K++ TLL+
Sbjct: 350 KKVIETLQDNYPEFVSRNIFINVPFWFYAMRAVLSPFLTQRTKSKFVVARPAKVRETLLK 409
Query: 250 EIDESQLPEIYGG 262
I +LP YGG
Sbjct: 410 YIPADELPVQYGG 422
>At4g39180 SEC14 - like protein
Length = 554
Score = 90.9 bits (224), Expect = 7e-19
Identities = 69/220 (31%), Positives = 106/220 (47%), Gaps = 19/220 (8%)
Query: 62 SKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQ 121
SK D M+ RFLRAR D++KA M+ + WRK + + + + K Y Q
Sbjct: 89 SKHDDHHMMLRFLRARKFDLEKAKQMWSDMLNWRKEYGADTIMEDFDFKEIEEVVKYYPQ 148
Query: 122 G---LDKKGRPIIVA----FAAKHFQNKNGLDAFKRYVVFALEKLISRMPPG-------- 166
G +DK+GRPI + A +D + +Y V EK + P
Sbjct: 149 GYHGVDKEGRPIYIERLGQVDATKLMKVTTIDRYVKYHVKEFEKTFNVKFPACSIAAKRH 208
Query: 167 EEKFVSIADIKGWGYANSD--IRGYLGALT-ILQDYYPERLGKLFIVHAPYMFMKVWKII 223
++ +I D++G G +N + + L ++ I D YPE L ++FI++A F +W +
Sbjct: 209 IDQSTTILDVQGVGLSNFNKAAKDLLQSIQKIDNDNYPETLNRMFIINAGCGFRLLWNTV 268
Query: 224 YPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGK 263
F+D T KI + N K + LLE ID ++LPE GGK
Sbjct: 269 KSFLDPKTTAKIHVLGN-KYQTKLLEIIDANELPEFLGGK 307
>At2g21540 putative phosphatidylinositol/phophatidylcholine transfer
protein
Length = 371
Score = 89.4 bits (220), Expect = 2e-18
Identities = 79/271 (29%), Positives = 121/271 (44%), Gaps = 21/271 (7%)
Query: 11 NSALDSKHELAKEETKGKILVQDDGALNDSTEAELTKINLMR-TLVESRDPSSKEVDDLM 69
N++ KH K T+ V ++D EL ++ R L+ SK D M
Sbjct: 37 NASNKFKHSFTKR-TRRNSRVMSVSIVDDIDLEELQAVDAFRQALILDELLPSKHDDHHM 95
Query: 70 IRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQ---GLDKK 126
+ RFLRAR D++KA M+ + WRK F + + + K Y Q G+DK
Sbjct: 96 MLRFLRARKFDLEKAKQMWTDMIHWRKEFGVDTIMEDFDFKEIDEVLKYYPQGYHGVDKD 155
Query: 127 GRPIIV----AFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPG--------EEKFVSIA 174
GRP+ + A +D + +Y V EK + P ++ +I
Sbjct: 156 GRPVYIERLGQVDATKLMQVTTIDRYVKYHVREFEKTFNIKLPACSIAAKKHIDQSTTIL 215
Query: 175 DIKGWGYA--NSDIRGYLGAL-TILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNT 231
D++G G + R L + I D YPE L ++FI++A F +W + F+D T
Sbjct: 216 DVQGVGLKSFSKAARDLLQRIQKIDSDNYPETLNRMFIINAGSGFRLLWSTVKSFLDPKT 275
Query: 232 KKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
KI + N K ++ LLE ID ++LPE GG
Sbjct: 276 TAKIHVLGN-KYQSKLLEIIDSNELPEFLGG 305
>At2g18180 putative phosphatidylinositol/phophatidylcholine transfer
protein
Length = 558
Score = 89.0 bits (219), Expect = 3e-18
Identities = 75/264 (28%), Positives = 121/264 (45%), Gaps = 25/264 (9%)
Query: 20 LAKEETKGKILVQDDGALNDSTEAELTKI--NLMRTLVESRDPSSKEVDDLMIRRFLRAR 77
L K+ K++ + D +AE K+ + L+ K D M+ RFL+AR
Sbjct: 32 LTKKRRSSKVMSVE--IFEDEHDAEELKVVDAFRQVLILDELLPDKHDDYHMMLRFLKAR 89
Query: 78 DLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG---LDKKGRPIIVAF 134
D++K + M+ ++WRK F + E + K Y QG +DK+GRP+ +
Sbjct: 90 KFDLEKTNQMWSDMLRWRKEFGADTVMEDFEFKEIDEVLKYYPQGHHGVDKEGRPVYIER 149
Query: 135 AAKHFQNK----NGLDAFKRYVVFALEKLISRMPPG--------EEKFVSIADIKGWGYA 182
+ K +D + Y V E+ + P ++ +I D++G G
Sbjct: 150 LGQVDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACSIAAKKHIDQSTTILDVQGVGLK 209
Query: 183 NSDIRGYLGALTILQ----DYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFV 238
N + + +T LQ D YPE L ++FI++A F +W + F+D T KI +
Sbjct: 210 NFN-KAARDLITRLQKVDGDNYPETLNRMFIINAGSGFRMLWNTVKSFLDPKTTAKIHVL 268
Query: 239 ENKKLKATLLEEIDESQLPEIYGG 262
N K ++ LLE ID S+LPE GG
Sbjct: 269 GN-KYQSKLLEIIDASELPEFLGG 291
>At2g16380 putative phosphatidylinositol/phosphatidylcholine
transfer protein
Length = 582
Score = 88.2 bits (217), Expect = 4e-18
Identities = 72/216 (33%), Positives = 104/216 (47%), Gaps = 20/216 (9%)
Query: 66 DDL-MIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQG-- 122
DDL M+ RFLRAR D +KA M+ ++WR F + E + K Y QG
Sbjct: 82 DDLHMMLRFLRARKFDKEKAKQMWSDMLQWRMDFGVDTIIEDFEFEEIDQVLKHYPQGYH 141
Query: 123 -LDKKGRPIIVAFAAKHFQNK----NGLDAFKRYVVFALEKLISRMPPG--------EEK 169
+DK+GRP+ + + NK +D +++Y V EK+ P ++
Sbjct: 142 GVDKEGRPVYIERLGQIDANKLLQATTMDRYEKYHVKEFEKMFKIKFPSCSAAAKKHIDQ 201
Query: 170 FVSIADIKGWGYAN--SDIRGYLGALT-ILQDYYPERLGKLFIVHAPYMFMKVWKIIYPF 226
+I D++G G N R L L I D YPE L ++FI++A F +W I F
Sbjct: 202 STTIFDVQGVGLKNFNKSARELLQRLLKIDNDNYPETLNRMFIINAGPGFRLLWAPIKKF 261
Query: 227 IDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
+D T KI + N K + LLE ID S+LP +GG
Sbjct: 262 LDPKTTSKIHVLGN-KYQPKLLEAIDASELPYFFGG 296
>At1g75370 sec14 cytosolic factor, putative
Length = 612
Score = 86.7 bits (213), Expect = 1e-17
Identities = 76/239 (31%), Positives = 109/239 (44%), Gaps = 22/239 (9%)
Query: 44 ELTKINLMRTLVESRDPSSKEVDDLMIR-RFLRARDLDVDKASAMFLKYMKWRKSFVPSG 102
EL ++ R L+ S + +DD I RFL+AR D+ K M+ +KWRK F
Sbjct: 87 ELRAVDEFRNLLVSENLLPPTLDDYHIMLRFLKARKFDIGKTKLMWSNMIKWRKDFGTDT 146
Query: 103 SVSPSEIADDLAQEKIY---VQGLDKKGRPIIV-----AFAAKHFQNKNGLDAFKRYVVF 154
E + K Y G+DK+GRP+ + AK Q ++ F RY V
Sbjct: 147 IFEDFEFEEFDEVLKYYPHGYHGVDKEGRPVYIERLGLVDPAKLMQVTT-VERFIRYHVR 205
Query: 155 ALEKLISRMPPG--------EEKFVSIADIKGWGYAN--SDIRGYLGALT-ILQDYYPER 203
EK ++ P + +I D++G G+ N R + L I D YPE
Sbjct: 206 EFEKTVNIKLPACCIAAKRHIDSSTTILDVQGVGFKNFSKPARDLIIQLQKIDNDNYPET 265
Query: 204 LGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
L ++FI++ F VW + F+D T KI + N K + LLE ID SQLP+ GG
Sbjct: 266 LHRMFIINGGSGFKLVWATVKQFLDPKTVTKIHVIGN-KYQNKLLEIIDASQLPDFLGG 323
>At2g21520 putative phosphatidylinositol/phosphatidylcholine
transfer protein
Length = 531
Score = 85.9 bits (211), Expect = 2e-17
Identities = 68/213 (31%), Positives = 101/213 (46%), Gaps = 21/213 (9%)
Query: 69 MIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQ---GLDK 125
M+ RFL+AR DV+KA M+ ++WRK F + + + K Y Q G+DK
Sbjct: 14 MMLRFLKARKFDVEKAKQMWADMIQWRKEFGTDTIIQDFDFEEINEVLKHYPQCYHGVDK 73
Query: 126 KGRPIIVAFAAKHFQNK----NGLDAFKRYVVFALEKLISRMPPG--------EEKFVSI 173
+GRPI + K N+ +D + RY V E+ P + +I
Sbjct: 74 EGRPIYIERLGKVDPNRLMQVTSMDRYVRYHVKEFERSFMIKFPSCTISAKRHIDSSTTI 133
Query: 174 ADIKGWGYANSDIRGYLGALTILQ----DYYPERLGKLFIVHAPYMFMKVWKIIYPFIDD 229
D++G G N + + +T LQ D YPE L ++FI++A F +W + F+D
Sbjct: 134 LDVQGVGLKNFN-KSARDLITRLQKIDGDNYPETLHQMFIINAGPGFRLLWNTVKSFLDP 192
Query: 230 NTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
T KI V K + LLE ID ++LPE GG
Sbjct: 193 KTSAKI-HVLGYKYLSKLLEVIDVNELPEFLGG 224
>At1g22180 unknown protein
Length = 249
Score = 83.6 bits (205), Expect = 1e-16
Identities = 54/180 (30%), Positives = 90/180 (50%), Gaps = 6/180 (3%)
Query: 87 MFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIY-VQGLDKKGRPIIVAFAAKHFQNKNGL 145
M + +KWR + P + EIA + KIY DK GR ++V + QN
Sbjct: 1 MLKETLKWRAQYKPE-EIRWEEIAREAETGKIYRANCTDKYGRTVLVMRPS--CQNTKSY 57
Query: 146 DAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLG 205
R +V+ +E I +P +E+ V + D G+ ++ ++ +LQ++YPERLG
Sbjct: 58 KGQIRILVYCMENAILNLPDNQEQMVWLIDFHGFNMSHISLKVSRETAHVLQEHYPERLG 117
Query: 206 KLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFV-ENKKLKATLLEEI-DESQLPEIYGGK 263
+ + P +F +K++ PF++ T K+ FV + L LLE++ D QL +GGK
Sbjct: 118 LAIVYNPPKIFESFYKMVKPFLEPKTSNKVKFVYSDDNLSNKLLEDLFDMEQLEVAFGGK 177
>At1g19650 sec14 cytosolic factor- like protein
Length = 608
Score = 82.8 bits (203), Expect = 2e-16
Identities = 74/238 (31%), Positives = 110/238 (46%), Gaps = 20/238 (8%)
Query: 44 ELTKINLMRTLVESRDPSSKEVDDLMIR-RFLRARDLDVDKASAMFLKYMKWRKSFVPSG 102
EL ++ R + S +DD I RFL AR D+ KA M+ ++WR+ F
Sbjct: 78 ELRYVSEFRQSLISDHLLPPNLDDYHIMLRFLFARKFDLGKAKLMWTNMIQWRRDFGTDT 137
Query: 103 SVSPSEIADDLAQEKIYVQG---LDKKGRPIIVAFAAKHFQNK----NGLDAFKRYVVFA 155
+ E + + Y QG +DK+GRP+ + K +K L+ + RY V
Sbjct: 138 ILEDFEFPELDEVLRYYPQGYHGVDKEGRPVYIERLGKVDASKLMQVTTLERYLRYHVKE 197
Query: 156 LEKLISRMPPG--------EEKFVSIADIKGWGYAN--SDIRGYLGALT-ILQDYYPERL 204
EK I+ P + +I D++G G N R + L I D YPE L
Sbjct: 198 FEKTITVKFPACCIAAKRHIDSSTTILDVQGLGLKNFTKTARDLIIQLQKIDSDNYPETL 257
Query: 205 GKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
++FI++A F +W + F+D T KI + N K + LLE ID SQLP+ +GG
Sbjct: 258 HRMFIINAGSGFKLLWGTVKSFLDPKTVSKIHVLGN-KYQNKLLEMIDASQLPDFFGG 314
>At3g22410 unknown protein
Length = 400
Score = 82.0 bits (201), Expect = 3e-16
Identities = 57/196 (29%), Positives = 96/196 (48%), Gaps = 8/196 (4%)
Query: 47 KINLMRTLVESRDP----SSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSG 102
K+ + LV+ + P K + + RFL+ + +V KA+ + WR++F
Sbjct: 7 KVEAVLRLVKKQSPLTFKQEKFCNRECVERFLKVKGDNVKKAAKQLSSCLSWRQNF-DIE 65
Query: 103 SVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKH-FQNKNGLDAFKRYVVFALEKLIS 161
+ E + +L+ Y+ G D++ RP+I+ F KH +Q + F R V F +E IS
Sbjct: 66 RLGAEEFSTELSDGVAYISGHDRESRPVII-FRFKHDYQKLHTQKQFTRLVAFTIETAIS 124
Query: 162 RMPPG-EEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVW 220
M E+ FV + D + +++ L L I+ D YP RL K FI+ P F +W
Sbjct: 125 SMSRNTEQSFVLLFDASFFRSSSAFANLLLATLKIIADNYPCRLYKAFIIDPPSFFSYLW 184
Query: 221 KIIYPFIDDNTKKKIV 236
K + PF++ +T I+
Sbjct: 185 KGVRPFVELSTATMIL 200
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,241,537
Number of Sequences: 26719
Number of extensions: 264917
Number of successful extensions: 821
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 722
Number of HSP's gapped (non-prelim): 46
length of query: 272
length of database: 11,318,596
effective HSP length: 98
effective length of query: 174
effective length of database: 8,700,134
effective search space: 1513823316
effective search space used: 1513823316
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC139708.6


