

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.14 - phase: 0
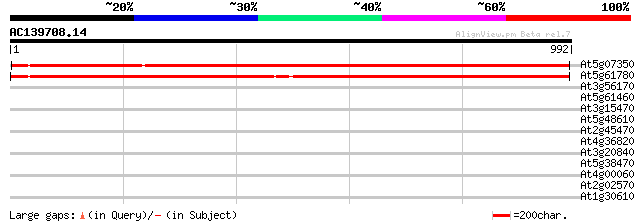
(992 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g07350 unknown protein 1405 0.0
At5g61780 100 kDa coactivator - like protein 1395 0.0
At3g56170 Ca(2+)-dependent nuclease 35 0.27
At5g61460 SMC-like protein (MIM) 33 0.80
At3g15470 unknown protein 32 1.4
At5g48610 putative protein 32 1.8
At2g45470 GPI-anchored protein (Fla8) 32 1.8
At4g36820 putative protein 32 2.3
At3g20840 putative transcription factor 32 2.3
At5g38470 DNA repair protein RAD23 homolog 30 6.8
At4g00060 unknown protein 30 6.8
At2g02570 unknown protein 30 6.8
At1g30610 hypothetical protein 30 8.8
>At5g07350 unknown protein
Length = 991
Score = 1405 bits (3636), Expect = 0.0
Identities = 712/996 (71%), Positives = 835/996 (83%), Gaps = 14/996 (1%)
Query: 3 ATAAGNSAWYKAKVKAVPSGDCIVVVSVAANAKLGVLPEKSITLSSLIAPRLARRGGVDE 62
AT A N W K +VKAV SGDC+V+ +++ N + G PEK+IT SSL+AP++ARRGG+DE
Sbjct: 2 ATGAENQ-WLKGRVKAVTSGDCLVITALSHN-RAGPPPEKTITFSSLMAPKMARRGGIDE 59
Query: 63 PFAWESREFLRKLLIGKEITFRIDYTVPSI-NREFGTVFLGDKNVALLVVSQGWAKVREQ 121
PFAWES+EFLRKL IGKE+ F++DY V +I REFG+VFLG++N+A LVV GWAKVRE
Sbjct: 60 PFAWESKEFLRKLCIGKEVAFKVDYKVEAIAGREFGSVFLGNENLAKLVVKTGWAKVREP 119
Query: 122 GQQ-KGEASPFLAELLRLEEQAKQEGLGRWSKVPGAAEASVRNLPPSALGDASNFDAMGL 180
GQQ + + SP++ ELL+LEE AKQEG GRWSKVPGAAEAS+RNLPPSA+GD++ FDAMGL
Sbjct: 120 GQQNQDKVSPYIKELLQLEELAKQEGYGRWSKVPGAAEASIRNLPPSAIGDSAGFDAMGL 179
Query: 181 LAKNKGVPMEALVEQVRDGSTLRIYLLPEFQFVQVFVAGIQAPQMGRRAAPESVVVPEVT 240
LA NKG PME +VEQVRDGST+R+YLLPEFQFVQVFVAG+QAP MGRR SVV E
Sbjct: 180 LAANKGKPMEGIVEQVRDGSTIRVYLLPEFQFVQVFVAGVQAPSMGRRTTNGSVV--ETV 237
Query: 241 VDTTNGDVPAEPRAPLTSAQRLAVSA-SAAETSADPFGADAKFFTEMRVLNRDVRIVLEG 299
D NGDV AE R PLT+AQRLA SA S+ E S+DPF +AK+FTE RVL+RDVRIVLEG
Sbjct: 238 PDEPNGDVSAESRGPLTTAQRLAASAASSVEVSSDPFATEAKYFTEHRVLSRDVRIVLEG 297
Query: 300 VDKFSNLIGSVYYPDGESAKDLALELVENGFAKYVEWSANMMEDEAKKKLKAAELEAKKT 359
VDKF+NLIGSV+Y DGE+ KDL LELVENG AK+VEWSANMME+EAKKKLKAAEL+ KK
Sbjct: 298 VDKFNNLIGSVHYSDGETVKDLGLELVENGLAKFVEWSANMMEEEAKKKLKAAELQCKKD 357
Query: 360 RLRIWTNYVPPTSNSKAIHDQNFTGKVVEVVSGDCVIVADDSIPYGSPQAERRVNLSSIR 419
++++W NYVPP +NSKAIHDQNFTGKVVEVVSGDC+IVADD++P+GSP AERRV LSSIR
Sbjct: 358 KVKMWANYVPPATNSKAIHDQNFTGKVVEVVSGDCLIVADDAVPFGSPAAERRVCLSSIR 417
Query: 420 CPKMGNPRRDEKPAPYAREAKEFLRTRLIGRQVNVQMEYSRKVGPVDGSAVPPGAVDSRV 479
PKMGNPRR+EKPAPYAREA+EFLR RLIG+QV VQMEYSRKV DG GA D R
Sbjct: 418 SPKMGNPRREEKPAPYAREAREFLRQRLIGKQVIVQMEYSRKVTQGDGPTTS-GAAD-RF 475
Query: 480 MDFGSVFVLSSGKADGDDAPSP---AVPASQQTGLNVAELIIGRGFGTVIRHRDFEERSN 536
MDFGSVF+ S+ KAD D+ +P A+ SQ G+N+AEL++ RGFG V+RHRDFEERSN
Sbjct: 476 MDFGSVFLPSAAKADSDEVTAPPAAAIAGSQPVGVNIAELVLVRGFGNVVRHRDFEERSN 535
Query: 537 FYDALLAAEARAISGRKGIHSAKDPPVMHITDLITASAKKAKDFLPFLHRSRRVPAVVEY 596
YDALLAAEARA++G+KGIHSAK+ P MHITDL ++AKKAKDFLP L R RR+PAVVEY
Sbjct: 536 HYDALLAAEARALAGKKGIHSAKESPAMHITDLTVSAAKKAKDFLPSLQRIRRIPAVVEY 595
Query: 597 VFSGHRFKLLIPKETCSIAFAFSGVRCPGREEPYSDEAIALMRRRIMQRDVEIEVETVDR 656
V SGHRFKL IPK TCSIAF+FSGVRCPGR EPYS+EAI++MRRRIMQRDVEIEVETVDR
Sbjct: 596 VLSGHRFKLYIPKITCSIAFSFSGVRCPGRGEPYSEEAISVMRRRIMQRDVEIEVETVDR 655
Query: 657 TGTFLGSLWESRANGAVPLLEAGLAKLQTSFGSDRIPDLHVLEQAEQSAKSKKLKIWENY 716
TGTFLGS+WESR N A LLEAGLAK+QTSFG+DRI + H+LEQAE+SAK++KLKIWENY
Sbjct: 656 TGTFLGSMWESRTNVATVLLEAGLAKMQTSFGADRIAEAHLLEQAERSAKNQKLKIWENY 715
Query: 717 VEGEVVPSGAN--VESKQQEVLKVTVTEVLGGGKFYVQTVGDQKIASIQNQLASLNLKDA 774
VEGE V +G VE++Q+E LKV VTEVLGGG+FYVQ+ GDQKIASIQNQLASL++KDA
Sbjct: 716 VEGEEVSNGNTNTVETRQKETLKVVVTEVLGGGRFYVQSAGDQKIASIQNQLASLSIKDA 775
Query: 775 PVIGAFNPKKGDIVLCYFHADSSWYRAMVVNTPRGPVESSKDAFEVFYIDYGNQEVVPYS 834
P+IG+FNPK+GDIVL F D+SW RAM+V PR V+S + FEVFYIDYGNQE VPYS
Sbjct: 776 PIIGSFNPKRGDIVLAQFSLDNSWNRAMIVTAPRAAVQSPDEKFEVFYIDYGNQETVPYS 835
Query: 835 QLRPLDPSVSAAPGLAQLCSLAYIKLPNLEEDFGQEAAEYLSELTLSSGKEFRAMVEEKD 894
+RP+DPSVSAAPGLAQLC LAYIK+P+LE+DFG EA EYL +TL SGKEF+A++EE+D
Sbjct: 836 AIRPIDPSVSAAPGLAQLCRLAYIKVPSLEDDFGPEAGEYLHTVTLGSGKEFKAVIEERD 895
Query: 895 TTGGKVKGQGTGPIIAVTLVAVDSEISVNAAMLQEGLARMEKRNRWDRTARKQALDNLEM 954
T+GGKVKGQGTG VTL+AVD EISVNAAMLQEG+ARMEKR +W ++ ALD LE
Sbjct: 896 TSGGKVKGQGTGTEFVVTLIAVDDEISVNAAMLQEGIARMEKRQKWGHKGKQAALDALEK 955
Query: 955 FQGEARTARRGMWQYGDIQSDDEDTAPPQRKAGGGR 990
FQ EAR +R G+WQYGDI+SDDEDT P ++ AGG R
Sbjct: 956 FQEEARKSRIGIWQYGDIESDDEDTGPARKPAGGRR 991
>At5g61780 100 kDa coactivator - like protein
Length = 985
Score = 1395 bits (3612), Expect = 0.0
Identities = 711/996 (71%), Positives = 831/996 (83%), Gaps = 18/996 (1%)
Query: 1 MAATAAGNSAWYKAKVKAVPSGDCIVVVSVAANAKLGVLPEKSITLSSLIAPRLARRGGV 60
MA AA + W K +VKAV SGDC+V+ ++ N + G PEK+ITLSSL+AP++ARRGG+
Sbjct: 1 MATGAATENQWLKGRVKAVTSGDCLVITALTHN-RAGPPPEKTITLSSLMAPKMARRGGI 59
Query: 61 DEPFAWESREFLRKLLIGKEITFRIDYTVPSI-NREFGTVFLGDKNVALLVVSQGWAKVR 119
DEPFAWESREFLRKL IGKE+ F++DY V +I REFG+V+LG++N+A LVV GWAKVR
Sbjct: 60 DEPFAWESREFLRKLCIGKEVAFKVDYKVEAIAGREFGSVYLGNENLAKLVVQNGWAKVR 119
Query: 120 EQGQQ-KGEASPFLAELLRLEEQAKQEGLGRWSKVPGAAEASVRNLPPSALGDASNFDAM 178
GQQ + + SP++AEL +LEEQA+QEG GRWSKVPGAAEAS+RNLPPSA+GD+ NFDAM
Sbjct: 120 RPGQQNQDKVSPYIAELEQLEEQAQQEGFGRWSKVPGAAEASIRNLPPSAVGDSGNFDAM 179
Query: 179 GLLAKNKGVPMEALVEQVRDGSTLRIYLLPEFQFVQVFVAGIQAPQMGRR-AAPESVVVP 237
GLLA +KG PME +VEQVRDGST+R+YLLPEFQFVQVFVAG+QAP MGRR + E+VV P
Sbjct: 180 GLLAASKGKPMEGIVEQVRDGSTIRVYLLPEFQFVQVFVAGLQAPSMGRRQSTQEAVVDP 239
Query: 238 EVTVDTTNGDVPAEPRAPLTSAQRLAVSA-SAAETSADPFGADAKFFTEMRVLNRDVRIV 296
+VT T+NGD AE R PLT+AQRLA SA S+ E S+DPF +AK+FTE+RVLNRDVRIV
Sbjct: 240 DVTA-TSNGDASAETRGPLTTAQRLAASAASSVEVSSDPFAMEAKYFTELRVLNRDVRIV 298
Query: 297 LEGVDKFSNLIGSVYYPDGESAKDLALELVENGFAKYVEWSANMMEDEAKKKLKAAELEA 356
LEGVDKF+NLIGSVYY DG++ KDL LELVENG AKYVEWSANM+++EAKKKLKA EL+
Sbjct: 299 LEGVDKFNNLIGSVYYSDGDTVKDLGLELVENGLAKYVEWSANMLDEEAKKKLKATELQC 358
Query: 357 KKTRLRIWTNYVPPTSNSKAIHDQNFTGKVVEVVSGDCVIVADDSIPYGSPQAERRVNLS 416
KK R+++W NYVPP SNSKAIHDQNFTGKVVEVVSGDC++VADDSIP+GSP AERRV LS
Sbjct: 359 KKNRVKMWANYVPPASNSKAIHDQNFTGKVVEVVSGDCLVVADDSIPFGSPMAERRVCLS 418
Query: 417 SIRCPKMGNPRRDEKPAPYAREAKEFLRTRLIGRQVNVQMEYSRKVGPVDGSAVPPGAVD 476
SIR PKMGNPRR+EKPAPYAREAKEFLR +LIG +V VQMEYSRK+ P DG V
Sbjct: 419 SIRSPKMGNPRREEKPAPYAREAKEFLRQKLIGMEVIVQMEYSRKISPGDG--VTTSGAG 476
Query: 477 SRVMDFGSVFVLSSGKADGDDAPSPAVPASQQTGLNVAELIIGRGFGTVIRHRDFEERSN 536
RVMDFGSVF+ S K D AV A+ G N+AELII RG GTV+RHRDFEERSN
Sbjct: 477 DRVMDFGSVFLPSPTKGD------TAVAAAATPGANIAELIISRGLGTVVRHRDFEERSN 530
Query: 537 FYDALLAAEARAISGRKGIHSAKDPPVMHITDLITASAKKAKDFLPFLHRSRRVPAVVEY 596
YDALLAAEARAI+G+K IHSAKD P +HI DL ASAKKAKDFLP L R ++ AVVEY
Sbjct: 531 HYDALLAAEARAIAGKKNIHSAKDSPALHIADLTVASAKKAKDFLPSLQRINQISAVVEY 590
Query: 597 VFSGHRFKLLIPKETCSIAFAFSGVRCPGREEPYSDEAIALMRRRIMQRDVEIEVETVDR 656
V SGHRFKL IPKE+CSIAFAFSGVRCPGR EPYS+EAIALMRR+IMQRDVEI VE VDR
Sbjct: 591 VLSGHRFKLYIPKESCSIAFAFSGVRCPGRGEPYSEEAIALMRRKIMQRDVEIVVENVDR 650
Query: 657 TGTFLGSLWE--SRANGAVPLLEAGLAKLQTSFGSDRIPDLHVLEQAEQSAKSKKLKIWE 714
TGTFLGS+WE S+ N LLEAGLAK+QT FG+DRIP+ H+LE AE+SAK++KLKIWE
Sbjct: 651 TGTFLGSMWEKNSKTNAGTYLLEAGLAKMQTGFGADRIPEAHILEMAERSAKNQKLKIWE 710
Query: 715 NYVEGEVVPSGAN-VESKQQEVLKVTVTEVLGGGKFYVQTVGDQKIASIQNQLASLNLKD 773
NYVEGE V +G++ VE++Q+E LKV VTEVLGGG+FYVQTVGDQK+ASIQNQLA+L+LKD
Sbjct: 711 NYVEGEEVVNGSSKVETRQKETLKVVVTEVLGGGRFYVQTVGDQKVASIQNQLAALSLKD 770
Query: 774 APVIGAFNPKKGDIVLCYFHADSSWYRAMVVNTPRGPVESSKDAFEVFYIDYGNQEVVPY 833
AP+IG+FNPKKGDIVL F D+SW RAM+VN PRG V+S ++ FEVFYIDYGNQE+VPY
Sbjct: 771 APIIGSFNPKKGDIVLAQFSLDNSWNRAMIVNGPRGAVQSPEEEFEVFYIDYGNQEIVPY 830
Query: 834 SQLRPLDPSVSAAPGLAQLCSLAYIKLPNLEEDFGQEAAEYLSELTLSSGKEFRAMVEEK 893
S +RP+DPSVS+APGLAQLC LAYIK+P EEDFG++A EYL +TL SGKEFRA+VEE+
Sbjct: 831 SAIRPVDPSVSSAPGLAQLCRLAYIKVPGKEEDFGRDAGEYLHTVTLESGKEFRAVVEER 890
Query: 894 DTTGGKVKGQGTGPIIAVTLVAVDSEISVNAAMLQEGLARMEKRNRWDRTARKQALDNLE 953
DT+GGKVKGQGTG + VTL+AVD EISVNAAMLQEG+ARMEKR RW+ ++ ALD LE
Sbjct: 891 DTSGGKVKGQGTGTELVVTLIAVDDEISVNAAMLQEGIARMEKRRRWEPKDKQAALDALE 950
Query: 954 MFQGEARTARRGMWQYGDIQSDDEDTAPPQRKAGGG 989
FQ EAR +R G+W+YGDIQSDDED P RK G G
Sbjct: 951 KFQDEARKSRTGIWEYGDIQSDDEDNV-PVRKPGRG 985
>At3g56170 Ca(2+)-dependent nuclease
Length = 323
Score = 34.7 bits (78), Expect = 0.27
Identities = 48/267 (17%), Positives = 110/267 (40%), Gaps = 27/267 (10%)
Query: 111 VSQGWAKVREQGQQKGEASPFLAELLRLEEQAKQEGLGRWSKVPGAAE-ASVRNLPPSAL 169
+ + W + + + + EAS + L+ ++A EGL + +P V P +L
Sbjct: 62 ILEAWKQAKPRPKTPEEASRLVIAALKNHQKADVEGLLSFYGLPSPHNLVEVPTEAPVSL 121
Query: 170 GDASNFDAMGLLAKNKGVPMEALVEQVRDGSTLRIYLLPEFQFVQ------VFVAGIQ-A 222
F+ L K V DG T+ +Y+ + V V +A ++ A
Sbjct: 122 PKGVRFELNTLPVDTKSVA---------DGDTVTVYVSSKDPLVSSSLPKDVSLAAVKRA 172
Query: 223 PQMGRRAAPESVVVPEVTVDTTNGDVPAEPRAPLTSAQRLAVSASAAETSADPFGADAKF 282
++ E+ + + + + + + L R+ +S + S P+G +A
Sbjct: 173 KAREKKNYTEADALHKTIIASGYRMISFQNEEVLAKKFRIRLSGIDSPESKMPYGKEAHD 232
Query: 283 FTEMRVLNRDVRIVLEGVDKFSNLIGSVYYPDGESAKDLALELVENGFAKYVEWSANMME 342
V + +++++ D++ +G +Y +G+ +++ L+ K + W + +
Sbjct: 233 ELLKMVEGKCLKVLVYTEDRYGRCVGDIYC-NGKFVQEVMLK-------KGLAW--HYVA 282
Query: 343 DEAKKKLKAAELEAKKTRLRIWTNYVP 369
+ + +L E EA++ R+ +W + P
Sbjct: 283 YDKRAELAKWENEARQKRVGLWASSNP 309
Score = 32.0 bits (71), Expect = 1.8
Identities = 36/169 (21%), Positives = 77/169 (45%), Gaps = 17/169 (10%)
Query: 549 ISGRKGIHSAKDPPVMHITDLITASAKKAKDFLPF--LHRSRRVPAVVEYVFSGHRFKLL 606
+S + + S+ P + + + A A++ K++ LH++ + SG+R
Sbjct: 149 VSSKDPLVSSSLPKDVSLAAVKRAKAREKKNYTEADALHKT--------IIASGYRMISF 200
Query: 607 IPKETCSIAFAF--SGVRCPGREEPYSDEAIALMRRRIMQRDVEIEVETVDRTGTFLGSL 664
+E + F SG+ P + PY EA + + + + +++ V T DR G +G +
Sbjct: 201 QNEEVLAKKFRIRLSGIDSPESKMPYGKEAHDELLKMVEGKCLKVLVYTEDRYGRCVGDI 260
Query: 665 WESRANGAVPLLEAGLAKLQTSFGSDRIPDLHVLEQAEQSAKSKKLKIW 713
+ + +L+ GLA ++ D+ + L + E A+ K++ +W
Sbjct: 261 YCNGKFVQEVMLKKGLAWHYVAY--DKRAE---LAKWENEARQKRVGLW 304
Score = 29.6 bits (65), Expect = 8.8
Identities = 26/103 (25%), Positives = 47/103 (45%), Gaps = 17/103 (16%)
Query: 56 RRGGVDEP-----FAWESREFLRKLLIGKEITFRIDYTVPSINREFGTVFLGDKNVALLV 110
R G+D P + E+ + L K++ GK + + YT R G ++ K V ++
Sbjct: 213 RLSGIDSPESKMPYGKEAHDELLKMVEGKCLKVLV-YTEDRYGRCVGDIYCNGKFVQEVM 271
Query: 111 VSQG--WAKVREQGQQKGEASPFLAELLRLEEQAKQEGLGRWS 151
+ +G W V + AEL + E +A+Q+ +G W+
Sbjct: 272 LKKGLAWHYVAYDKR---------AELAKWENEARQKRVGLWA 305
>At5g61460 SMC-like protein (MIM)
Length = 1057
Score = 33.1 bits (74), Expect = 0.80
Identities = 27/101 (26%), Positives = 51/101 (49%), Gaps = 9/101 (8%)
Query: 643 MQRDVEIEVETVDRTGTFLGSLW------ESRANGAVPLLE--AGLAKLQTSFGSDRIPD 694
+QR++ ++E +D FL L E +AN L E AK + + +
Sbjct: 718 LQREIMKDLEEIDEKEAFLEKLQNCLKEAELKANKLTALFENMRESAKGEIDAFEEAENE 777
Query: 695 LHVLEQAEQSAKSKKLKIWENYVEGEVVPSGANVESKQQEV 735
L +E+ QSA+++K+ +EN ++ +V+P N E+ +E+
Sbjct: 778 LKKIEKDLQSAEAEKIH-YENIMKNKVLPDIKNAEANYEEL 817
>At3g15470 unknown protein
Length = 883
Score = 32.3 bits (72), Expect = 1.4
Identities = 27/106 (25%), Positives = 46/106 (42%), Gaps = 5/106 (4%)
Query: 863 LEEDFGQEAAEYLSELTLSSGKEFRAMVEEKDTTGGKVKGQGTGPIIAVTLVAVDSEISV 922
L E E E + L +GKEF ++D T KVK GTG + + E+ V
Sbjct: 234 LRESVVNEEVEVCTIKNLDNGKEFVVNEIQEDGTWKKVKEVGTG----TQMTMEEFEMCV 289
Query: 923 NAAMLQEGLARMEKRNRWDRTARKQALDNLEMFQGEA-RTARRGMW 967
+ + + L R + D+ K+ D+ + A ++ ++G W
Sbjct: 290 GHSPIVQELMRRQNVEDSDKNTSKENEDSGNSNKDNASKSKKKGSW 335
>At5g48610 putative protein
Length = 470
Score = 32.0 bits (71), Expect = 1.8
Identities = 46/184 (25%), Positives = 74/184 (40%), Gaps = 24/184 (13%)
Query: 127 EASPFLAELLRLEEQAKQEGLGRWSKVPGAAEASVRNLPPSA-LGDASNFDAMGLLAKNK 185
+ P L +LEE+ K R K+P + S++NL LG + G L +N
Sbjct: 251 QEKPKLIRGPKLEEREKDSPDLRNCKLPDVSRTSIKNLHTEGNLGKRKDHMTNGFLYENG 310
Query: 186 GVP-----MEALVEQVRDGSTLRIYLLPEFQFVQVFVAGIQAPQMGRRAAPESVVVPEVT 240
P + A V V +G TL P +V G P+ V EV
Sbjct: 311 TTPHKLQKLSASVPSVENGRTLGAPRTPPMPASEV---------QGTTCKPQ---VKEVR 358
Query: 241 VD--TTNGDV-PAEPRAPLTSAQRLAVSASAAETSADPFGADAKFFTEMRVLNRDVRIVL 297
++ +G+ P +PL + ++ V + E SA P D K+ + +L+ R +L
Sbjct: 359 INGFAVSGEKRKVCPPSPLAATMKVKVKEN-GEASAKPPHPDLKYLNQ--ILHVPTRELL 415
Query: 298 EGVD 301
+D
Sbjct: 416 PEID 419
>At2g45470 GPI-anchored protein (Fla8)
Length = 420
Score = 32.0 bits (71), Expect = 1.8
Identities = 46/199 (23%), Positives = 72/199 (36%), Gaps = 23/199 (11%)
Query: 102 GDKNVALLVVSQGWAKVREQGQQKG-------------EASPFLAELLRLEEQAKQE--G 146
G K A L+VS G K E +KG E P L +L + E + E
Sbjct: 198 GCKTFANLLVSSGVLKTYESAVEKGLTVFAPSDEAFKAEGVPDLTKLTQAEVVSLLEYHA 257
Query: 147 LGRWSKVPGAAEASVRNLPPSALGDASNFDAMGLLAKNK-----GVPMEALVEQVRDGST 201
L + K G+ + + N+ A A FD + ++ GV L + V D +
Sbjct: 258 LAEY-KPKGSLKTNKNNISTLATNGAGKFDLTTSTSGDEVILHTGVAPSRLADTVLDATP 316
Query: 202 LRIYLLPEFQFVQVFVAGIQAPQMGRRAAPESVVVPEVTVDTTNGDVPAEPRAPLTSAQR 261
+ I+ + ++P APE V P + A P AP T
Sbjct: 317 VVIFTVDNVLLPAELFGKSKSPSPA--PAPEPVTAPTPSPADAPSPTAASPPAPPTDESP 374
Query: 262 LAVSASAAETSADPFGADA 280
+ + + SA+ A+A
Sbjct: 375 ESAPSDSPTGSANSKSANA 393
>At4g36820 putative protein
Length = 344
Score = 31.6 bits (70), Expect = 2.3
Identities = 33/120 (27%), Positives = 48/120 (39%), Gaps = 22/120 (18%)
Query: 259 AQRLAVSASAAETSADPFGADAKFFTEMRVLNRDVRIVLEGVDKFSNLIGSVYYPDGESA 318
A R V E + DP D R LN+ + G+D S + GES
Sbjct: 35 AMRTRVPKDIGEATIDPEPGDLTI--SQRFLNK---FSMNGIDTTSKM------SIGESL 83
Query: 319 KDLALELVEN-------GFAKYVEWSANMMEDEAKKKLKAAELEAKKTRL----RIWTNY 367
+ E+ N G + E + + + KK L+AAE+E KT+L +IW Y
Sbjct: 84 MEKLKEMDMNKDRIRLDGLSHPKEETLGLTVQDVKKLLRAAEIEVIKTKLMETGKIWIRY 143
>At3g20840 putative transcription factor
Length = 529
Score = 31.6 bits (70), Expect = 2.3
Identities = 28/112 (25%), Positives = 45/112 (40%), Gaps = 1/112 (0%)
Query: 869 QEAAEYLSELTLSSGKEFRAMVEEKDTTGGKVKGQGTGPIIAVTLVAVDSEISVNAAMLQ 928
QE+A L LTLS G V +K + +G +AV A +++ +
Sbjct: 78 QESAHNLQSLTLSMGTTAGNNVVDKASPSETTGDNASGGALAVVETATPRR-ALDTFGQR 136
Query: 929 EGLARMEKRNRWDRTARKQALDNLEMFQGEARTARRGMWQYGDIQSDDEDTA 980
+ R R+RW DN +G++R R+G + D + D A
Sbjct: 137 TSIYRGVTRHRWTGRYEAHLWDNSCRREGQSRKGRQGGYDKEDKAARSYDLA 188
>At5g38470 DNA repair protein RAD23 homolog
Length = 378
Score = 30.0 bits (66), Expect = 6.8
Identities = 26/112 (23%), Positives = 45/112 (39%), Gaps = 15/112 (13%)
Query: 196 VRDGSTLRIYLLPEFQFVQVFVAGIQAPQMGRRAA---------PESVVVPEVTVDTTNG 246
++D +TL + E F+ + ++ +A G A P++V P+V+ T +
Sbjct: 52 LKDETTLEENNVVENSFIVIMLSKTKASPSGASTASAPAPSATQPQTVATPQVSAPTASV 111
Query: 247 DVPAEPRAPLTSAQRLAVSASAAETSADPFGADAKFFTEMRVLNRDVRIVLE 298
VP A A A+AA D +G A L V+ +L+
Sbjct: 112 PVPTSGTA------TAAAPATAASVQTDVYGQAASNLVAGTTLESTVQQILD 157
>At4g00060 unknown protein
Length = 1344
Score = 30.0 bits (66), Expect = 6.8
Identities = 17/73 (23%), Positives = 32/73 (43%)
Query: 399 DDSIPYGSPQAERRVNLSSIRCPKMGNPRRDEKPAPYAREAKEFLRTRLIGRQVNVQMEY 458
DD+ SP + L S + G R +KP P A+ K + G++ ++E+
Sbjct: 270 DDNTHKCSPSSSSNQKLGSTNRKQKGKTRNMKKPTPEAKSDKNVNLSTKNGKKDQAKLEF 329
Query: 459 SRKVGPVDGSAVP 471
++ ++ VP
Sbjct: 330 NKSREAIECKKVP 342
>At2g02570 unknown protein
Length = 300
Score = 30.0 bits (66), Expect = 6.8
Identities = 34/136 (25%), Positives = 61/136 (44%), Gaps = 20/136 (14%)
Query: 726 ANVESKQQEVLKVTVTEVLGGGKFYVQTVGDQKIAS--------IQNQLASLNLKDAPVI 777
A++E + +EV+ +T EVL K ++ D +++ ++ L++ P+
Sbjct: 41 ADMEKELKEVIALT-EEVLATAKQNEISLSDAGVSAEATPGSPDLEGAWEKTGLRNDPIH 99
Query: 778 GAFNPKKGDIVLCYFHADSSWYRAMV-VNTPRGPVESSKDAFEVFYIDYGNQEVVPYSQL 836
P G V F D WY A + +T G + V Y ++GN+E V +
Sbjct: 100 EGKFPV-GTKVQAVFSDDGEWYDATIEAHTANG--------YFVAYDEWGNKEEVDPDNV 150
Query: 837 RPLDP-SVSAAPGLAQ 851
RP++ ++ A LAQ
Sbjct: 151 RPIEQNAIVEAERLAQ 166
>At1g30610 hypothetical protein
Length = 1006
Score = 29.6 bits (65), Expect = 8.8
Identities = 23/81 (28%), Positives = 39/81 (47%), Gaps = 2/81 (2%)
Query: 282 FFTEMRVLNRDVRIVLEGVDKFSNLIGSVY-YPDGESAKDLALELVENGFAKYVEWSANM 340
+ T + VL + R V E ++ F ++ + YPD + + +A+ L + G K + + +
Sbjct: 510 YTTALNVLGKSRRPV-EALNVFHAMLLQISSYPDMVAYRSIAVTLGQAGHIKELFYVIDT 568
Query: 341 MEDEAKKKLKAAELEAKKTRL 361
M KKK K LE RL
Sbjct: 569 MRSPPKKKFKPTTLEKWDPRL 589
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,745,843
Number of Sequences: 26719
Number of extensions: 937441
Number of successful extensions: 2466
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 2421
Number of HSP's gapped (non-prelim): 17
length of query: 992
length of database: 11,318,596
effective HSP length: 109
effective length of query: 883
effective length of database: 8,406,225
effective search space: 7422696675
effective search space used: 7422696675
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC139708.14


