

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.8 - phase: 0
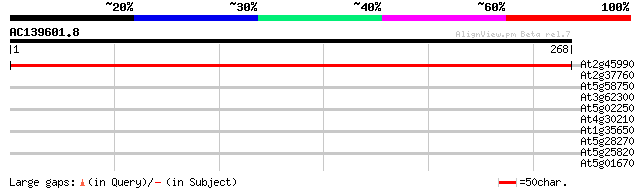
(268 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g45990 unknown protein 427 e-120
At2g37760 putative alcohol dehydrogenase 30 1.8
At5g58750 wounding stress induced protein - like 29 2.3
At3g62300 putative protein 28 3.9
At5g02250 ribonuclease II-like protein 28 6.7
At4g30210 NADPH-ferrihemoprotein reductase (ATR2) 28 6.7
At1g35650 hypothetical protein 28 6.7
At5g28270 putative protein 27 8.7
At5g25820 putative protein 27 8.7
At5g01670 aldose reductase-like protein 27 8.7
>At2g45990 unknown protein
Length = 268
Score = 427 bits (1097), Expect = e-120
Identities = 203/268 (75%), Positives = 235/268 (86%)
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
MG LYALDFDGVLCD+CGE+++SA+KAAK+RWPDLF VDS+ E+WIV+QM VRPVVET
Sbjct: 1 MGDLYALDFDGVLCDSCGESSLSAVKAAKVRWPDLFEGVDSALEEWIVDQMHIVRPVVET 60
Query: 61 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFG 120
GYE LLLVRLLLET++PSIRKSSVAEGLTV+GILE W K KP++ME W+E+RD L+DLFG
Sbjct: 61 GYENLLLVRLLLETKIPSIRKSSVAEGLTVDGILESWAKFKPVIMEAWDEDRDALVDLFG 120
Query: 121 KVRDDWLENDFAGWIQGNRFYPGVADALRFASSKVYIVTTKQGRFADALLRELAGITIPP 180
KVRDDW+ D WI NRFYPGV+DAL+FASSK+YIVTTKQGRFA+ALLRE+AG+ IP
Sbjct: 121 KVRDDWINKDLTTWIGANRFYPGVSDALKFASSKIYIVTTKQGRFAEALLREIAGVIIPS 180
Query: 181 ERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNWGF 240
ERIYGLG+GPKVE LK LQ PEHQGLTLHFVEDR+A LKNVIKEPELD W+LYL WG+
Sbjct: 181 ERIYGLGSGPKVEVLKLLQDKPEHQGLTLHFVEDRLATLKNVIKEPELDKWSLYLGTWGY 240
Query: 241 NTQKERDEAAANPRIQLIDLSDFSSKLK 268
NT+KER EAA PRIQ+I+LS FS+KLK
Sbjct: 241 NTEKERAEAAGIPRIQVIELSTFSNKLK 268
>At2g37760 putative alcohol dehydrogenase
Length = 311
Score = 29.6 bits (65), Expect = 1.8
Identities = 16/59 (27%), Positives = 32/59 (54%), Gaps = 4/59 (6%)
Query: 210 HFVEDRIAALKNVIKEPELDNWNLYLVNWGFNTQKERDEAAANPRIQLIDLSDFSSKLK 268
H ED AL+ +++ ++D +LYL++W + +KE + P +++ D +S K
Sbjct: 83 HLPEDVPKALEKTLQDLQIDYVDLYLIHWPASLKKE----SLMPTPEMLTKPDITSTWK 137
>At5g58750 wounding stress induced protein - like
Length = 386
Score = 29.3 bits (64), Expect = 2.3
Identities = 16/84 (19%), Positives = 36/84 (42%), Gaps = 7/84 (8%)
Query: 179 PPERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKEPELDNWNLYLVNW 238
P RIYG+ P++ ++ K+ L + R++ L++++ +++ V W
Sbjct: 41 PGWRIYGVARNPEINSMTKMYNFISCDLLNASETKQRLSPLQDIVS-------HVFWVTW 93
Query: 239 GFNTQKERDEAAANPRIQLIDLSD 262
+ DE + L++ D
Sbjct: 94 SGEFPLDTDECCVQNKTMLMNALD 117
>At3g62300 putative protein
Length = 708
Score = 28.5 bits (62), Expect = 3.9
Identities = 18/61 (29%), Positives = 28/61 (45%)
Query: 166 ADALLRELAGITIPPERIYGLGTGPKVETLKKLQKMPEHQGLTLHFVEDRIAALKNVIKE 225
AD ++ E A +T PE + T K KKLQ M + T V ++++ K +
Sbjct: 385 ADVVMNESAPVTSQPEIAAPVKTQGKTTPKKKLQAMKNQKSSTNDSVGEKVSVNKRKRGQ 444
Query: 226 P 226
P
Sbjct: 445 P 445
>At5g02250 ribonuclease II-like protein
Length = 803
Score = 27.7 bits (60), Expect = 6.7
Identities = 12/28 (42%), Positives = 20/28 (70%)
Query: 192 VETLKKLQKMPEHQGLTLHFVEDRIAAL 219
+E L++ +K ++ L L FV+DRIA+L
Sbjct: 733 IEFLRRQEKGKKYTALVLRFVKDRIASL 760
>At4g30210 NADPH-ferrihemoprotein reductase (ATR2)
Length = 711
Score = 27.7 bits (60), Expect = 6.7
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query: 8 DFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTED 45
D GVLCD ET AL+ + PD + S+ + ED
Sbjct: 350 DHVGVLCDNLSETVDEALRLLDMS-PDTYFSLHAEKED 386
>At1g35650 hypothetical protein
Length = 622
Score = 27.7 bits (60), Expect = 6.7
Identities = 27/121 (22%), Positives = 53/121 (43%), Gaps = 12/121 (9%)
Query: 10 DGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVETGYETLLLVR 69
DGVLC T G + IS + D+F + T + ++ ++ V G + LL
Sbjct: 168 DGVLCPTSGRSKISPVHVDMAENLDMFLNFPWGTMSF----LMTLKSVTARGADKLLRKS 223
Query: 70 LLLETRVPSIRKSSVAEGLTVEGILEDWFKLKPIVMEEWNENRDDLIDLFGKVRDDWLEN 129
+ ++ VP + + + G++ D + +E N+N D+ + V +D ++
Sbjct: 224 VTVQV-VPVLSGTPTGDAFKSGGVVPD------SLPDERNDN-SDVCSTYDCVDEDTIQP 275
Query: 130 D 130
D
Sbjct: 276 D 276
>At5g28270 putative protein
Length = 526
Score = 27.3 bits (59), Expect = 8.7
Identities = 24/94 (25%), Positives = 42/94 (44%), Gaps = 18/94 (19%)
Query: 1 MGHLYALDFDGVLCDTCGETAISALKAAKLRWPDLFGSVDSSTEDWIVEQMIKVRPVVET 60
+ + L+ + L ++ ET + K W LFGS +++ P VE
Sbjct: 132 LARITGLECETYLKNSAIETVMQRKAGEKTVWNTLFGSANAN-------------PKVED 178
Query: 61 GYETLLLVRLLLETRVPSIRKSSVAEGLTVEGIL 94
L RL+ ET +P +K ++A + V+G+L
Sbjct: 179 -----LRDRLISETDMPPWKKLALALIIIVDGVL 207
>At5g25820 putative protein
Length = 654
Score = 27.3 bits (59), Expect = 8.7
Identities = 9/23 (39%), Positives = 16/23 (69%)
Query: 100 LKPIVMEEWNENRDDLIDLFGKV 122
L+PI++ W N+D + +FGK+
Sbjct: 503 LRPILLSYWGNNKDPDLKIFGKL 525
>At5g01670 aldose reductase-like protein
Length = 322
Score = 27.3 bits (59), Expect = 8.7
Identities = 10/21 (47%), Positives = 17/21 (80%)
Query: 218 ALKNVIKEPELDNWNLYLVNW 238
AL+N +KE +L+ +LYL++W
Sbjct: 102 ALQNTLKELQLEYLDLYLIHW 122
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,075,750
Number of Sequences: 26719
Number of extensions: 256467
Number of successful extensions: 611
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 605
Number of HSP's gapped (non-prelim): 10
length of query: 268
length of database: 11,318,596
effective HSP length: 98
effective length of query: 170
effective length of database: 8,700,134
effective search space: 1479022780
effective search space used: 1479022780
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC139601.8


