

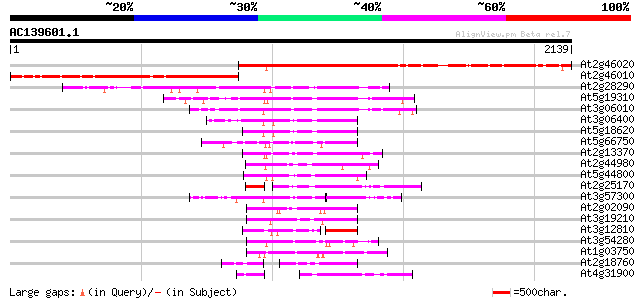
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.1 - phase: 0 /pseudo
(2139 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g46020 putative SNF2 subfamily transcriptional activator 1485 0.0
At2g46010 hypothetical protein 804 0.0
At2g28290 putative SNF2 subfamily transcription regulator 439 e-123
At5g19310 homeotic gene regulator - like protein 439 e-122
At3g06010 putative transcriptional regulator 437 e-122
At3g06400 putative ATPase (ISW2-like) 320 7e-87
At5g18620 chromatin remodelling complex ATPase chain ISWI -like ... 313 9e-85
At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1) 272 1e-72
At2g13370 pseudogene 271 2e-72
At2g44980 putative transcription activator 221 5e-57
At5g44800 helicase-like protein 217 7e-56
At2g25170 putative chromodomain-helicase-DNA-binding protein 201 5e-51
At3g57300 helicase-like protein 194 5e-49
At2g02090 putative helicase 188 3e-47
At3g19210 DNA repair protein, putative 177 5e-44
At3g12810 Snf2-related CBP activator protein, putative 177 6e-44
At3g54280 TATA box binding protein (TBP) associated factor (TAF)... 168 3e-41
At1g03750 hypothetical protein 168 4e-41
At2g18760 putative SNF2/RAD54 family DNA repair and recombinatio... 147 9e-35
At4g31900 putative protein 145 3e-34
>At2g46020 putative SNF2 subfamily transcriptional activator
Length = 1245
Score = 1485 bits (3844), Expect = 0.0
Identities = 814/1310 (62%), Positives = 966/1310 (73%), Gaps = 116/1310 (8%)
Query: 874 SVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKT 933
S S+YY LAHAVNE V+RQPSML+AGTLR+YQLVGLQWMLSLYNNKLNGILADEMGLGKT
Sbjct: 7 SNSRYYTLAHAVNEVVVRQPSMLQAGTLRDYQLVGLQWMLSLYNNKLNGILADEMGLGKT 66
Query: 934 VQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFNP----ENCI-------------D 976
VQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWK + +CI
Sbjct: 67 VQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKSELHTWLPSVSCIYYVGTKDQRSKLFS 126
Query: 977 HAE*IAYMVTIGVMHFLCWIEGSSVK------IIFSTQRMKDRESVLARDLDRYRCHRRL 1030
+ + V + F+ + K II QRMKDRESVLARDLDRYRC RRL
Sbjct: 127 QVKFEKFNVLVTTYEFIMYDRSKLSKVDWKYIIIDEAQRMKDRESVLARDLDRYRCQRRL 186
Query: 1031 LLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKV 1090
LLTGTPLQNDLKELWSLLNLLLP+VFDN+KAF+DWF++PFQKE P N E+DWLETEKKV
Sbjct: 187 LLTGTPLQNDLKELWSLLNLLLPDVFDNRKAFHDWFAQPFQKEGPAHNIEDDWLETEKKV 246
Query: 1091 IIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPE 1150
I+IHRLHQILEPFMLRRRVE+VEGSLP KVS+VLRCRMSA QSA+YDWIK+TGTLR++P+
Sbjct: 247 IVIHRLHQILEPFMLRRRVEDVEGSLPAKVSVVLRCRMSAIQSAVYDWIKATGTLRVDPD 306
Query: 1151 EEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSDLSKDFMVKCCGKLWMLD 1210
+E+ R +K+P+YQAK Y+TLNNRCMELRK CNHPLLNYP+F+D SKDF+V+ CGKLW+LD
Sbjct: 307 DEKLRAQKNPIYQAKIYRTLNNRCMELRKACNHPLLNYPYFNDFSKDFLVRSCGKLWILD 366
Query: 1211 RILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPN 1270
RILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTT+LEDRESAIVDFN P+
Sbjct: 367 RILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTSLEDRESAIVDFNDPD 426
Query: 1271 SDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEA 1330
+DCFIFLLSIRAAGRGLNLQ+ADTVVIYDPDPNPKNEEQAVARAHRIGQ REVKVIYMEA
Sbjct: 427 TDCFIFLLSIRAAGRGLNLQTADTVVIYDPDPNPKNEEQAVARAHRIGQTREVKVIYMEA 486
Query: 1331 VVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGR 1390
VV+K+SSHQKEDE+R GG++D+ED++AGKDRYIGSIE LIR+NIQQYKIDMADEVINAGR
Sbjct: 487 VVEKLSSHQKEDELRSGGSVDLEDDMAGKDRYIGSIEGLIRNNIQQYKIDMADEVINAGR 546
Query: 1391 FDQRTTHEERRLTLETLLHDEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEEEDW 1450
FDQRTTHEERR+TLETLLHDEER QETVHDVPSL EVNRMIAR+EEEVELFDQMDEE DW
Sbjct: 547 FDQRTTHEERRMTLETLLHDEERYQETVHDVPSLHEVNRMIARSEEEVELFDQMDEEFDW 606
Query: 1451 LEEMTRYDQVPDWIRASTREVNAAIAASSKRPSKKNALSGGNVVLDSTEIGSERRRGRPK 1510
EEMT ++QVP W+RASTREVNA +A SK+PS KN LS N+++ G ER+RGRPK
Sbjct: 607 TEEMTNHEQVPKWLRASTREVNATVADLSKKPS-KNMLSSSNLIVQPGGPGGERKRGRPK 665
Query: 1511 GKKNPSYKELEDS----SEEISEDRNEDSAH-DEGEIGEFEDDGYSGAGIAQPVDKDKLD 1565
KK +YKE+ED SEE SE+RN DS + +EG+I +F+DD +GA +K + D
Sbjct: 666 SKK-INYKEIEDDIAGYSEESSEERNIDSGNEEEGDIRQFDDDELTGALGDHQTNKGEFD 724
Query: 1566 DVTPSDAEYECPRSSSESARNNNVVEGGSSASSAGVQRLTQAVSPSVSSQKFASLSALDA 1625
P Y+ P S +N + GSS SS R + SP VSSQKF SLSALD
Sbjct: 725 GENPV-CGYDYPPGSGSYKKNPPRDDAGSSGSSPESHRSKEMASP-VSSQKFGSLSALDT 782
Query: 1626 KPSSISKKMVHGFILYFEYFILLPGGFYSIAFLLISSDKIYQGDELEEGEIAVSGESHMY 1685
+P S+SK+++ D+LEEGEIA SG+SH+
Sbjct: 783 RPGSVSKRLL---------------------------------DDLEEGEIAASGDSHID 809
Query: 1686 HQQSGSWIHDRDEGEEEQVLQKPKIKRKRSLRVRPRHTMEKPEDKSGSEMASLQRGQSFL 1745
Q+SGSW HDRDEG+EEQVLQ P IKRKRS+R+RPR T E+ + GSEM + Q L
Sbjct: 810 LQRSGSWAHDRDEGDEEQVLQ-PTIKRKRSIRLRPRQTAERVD---GSEMPAAQP----L 861
Query: 1746 LPDKKYPLQSRINQESKTFGDSSSNKHDKNEPILKNKRNLPARKVANASKLHV-SPKSSR 1804
D+ Y + +T DS S++ D+++ R++PA+KVA+ SKLHV SPKS R
Sbjct: 862 QVDRSY------RSKLRTVVDSHSSRQDQSDS-SSRLRSVPAKKVASTSKLHVSSPKSGR 914
Query: 1805 LNCTSAPSEDNDEHSRERLKGKPNNLRGSSAHVTNMTEIIQRRCKSVISKLQRRIDKEGH 1864
LN T EDN E SRE G + SS M+ IIQ+RCK VISKLQRRIDKEG
Sbjct: 915 LNATQLTVEDNAEASRETWDG--TSPISSSNAGARMSHIIQKRCKIVISKLQRRIDKEGQ 972
Query: 1865 QIVPLLTDLWKRIENSGFAGGSGNNLLDLRKIDQRINRLEYSGVMEFVFDVQFMLKSAMQ 1924
QIVP+LT+LWKRI+N G+A G NNLL+LR+ID R+ RLEY+GVME DVQ ML+ AMQ
Sbjct: 973 QIVPMLTNLWKRIQN-GYAAGGVNNLLELREIDHRVERLEYAGVMELASDVQLMLRGAMQ 1031
Query: 1925 FYGYSYEVRTEARKVHDLFFDILKTTFSDIDFGEAKSALSFT-SQISANAGASSKQATVF 1983
FYG+S+EVR+EA+KVH+LFFD+LK +F D DF EA++ALSF+ S + + + + A +
Sbjct: 1032 FYGFSHEVRSEAKKVHNLFFDLLKMSFPDTDFREARNALSFSGSAPTLVSTPTPRGAGIS 1091
Query: 1984 PSKRKRGKNDMETDPTPTQKPLQRGSTSNSESGRIKVQLPQKASRTGSGSGSAREQLQQD 2043
KR++ N+ ET+P+ Q+ QR E+ RI+VQ+PQK ++ G + D
Sbjct: 1092 QGKRQKLVNEPETEPSSPQRSQQR------ENSRIRVQIPQKETKLGGTTS------HTD 1139
Query: 2044 SPSLLTHPGDLVVCKKKRNERGDKSSVKHRI-GSAGPVSPPKIVVHTVLAERSPTPGSGS 2102
+L HPG+LV+CKKKR +R +KS K R GS+ PVSPP ++ L RSP SG
Sbjct: 1140 ESPILAHPGELVICKKKRKDR-EKSGPKTRTGGSSSPVSPPPAMIGRGL--RSPV--SGG 1194
Query: 2103 TPR-------------AGHAHTSNGSGGSVGWANPVKRMRTDSGKRRPSH 2139
PR H + S +G SVGWANPVKR+RTDSGKRRPSH
Sbjct: 1195 VPRETRLAQQQRWPNQPTHPNNSGAAGDSVGWANPVKRLRTDSGKRRPSH 1244
>At2g46010 hypothetical protein
Length = 942
Score = 804 bits (2076), Expect = 0.0
Identities = 478/890 (53%), Positives = 593/890 (65%), Gaps = 48/890 (5%)
Query: 1 MQLPPQSRNFFALAQHGPNQGQGIE-----QQRLNPVRQAYSQYALQSFQQRPALAMQSQ 55
MQ+P QSRNFF Q Q Q QQ NP++QAY Q+A+Q+ Q+ +Q
Sbjct: 81 MQMPQQSRNFFESPQQQQQQQQQGSSTQEGQQNFNPMQQAYIQFAMQAQHQK------AQ 134
Query: 56 QQPKMEMLGPTSV-KDQEMRMGNFKLQDLMSMQAVNHGQGSSSSRNSSEHFSHGEKRVEQ 114
QQ +M M+G +SV KDQ+ RMG +QDL +SSS+ S + F+ GE++ E
Sbjct: 135 QQARMGMVGSSSVGKDQDARMGMLNMQDLNPSSQPQ----ASSSKPSGDQFARGERQTES 190
Query: 115 GQQLASDKKNEGKSSTQG-LGIGHLMPGNNIRPVQALPTQQSIPIAMNNQIATSDQLRAM 173
Q ++NE KS Q +G G LMPGN IRP+QA QQ + NNQ+A + Q +AM
Sbjct: 191 SSQ----QRNETKSHPQQQVGTGQLMPGNMIRPMQAPQAQQLVNNMGNNQLAFAQQWQAM 246
Query: 174 QAWAHERNIDLSQPANANFAAQLNLMQTRMVQQSKES----GAQSSSVPVSKQQATSPAV 229
QAWA ERNIDLS PANA+ A +++Q RM Q K +QS S+P+S Q A+S V
Sbjct: 247 QAWARERNIDLSHPANASQMA--HILQARMAAQQKAGEGNVASQSPSIPISSQPASSSVV 304
Query: 230 SSEGSAHANSSTDVSALVGSVKARQTAPPSHLGLPINAGVAGNSSDTAVQQFSLHGRDAQ 289
E S HANS++D+S GS KAR + + + A+ FS GR+
Sbjct: 305 PGENSPHANSASDISGQSGSAKARHALSTGSFASTSSPRMV----NPAMNPFSGQGRENP 360
Query: 290 GSLKQLIVGVNGMPSMHPQQSSANKSLGADSSLNAKASSSRSDPEPAKMQYVRQLSQHAS 349
+ L+ NGMPS +P Q+SAN++ D NA S E +MQ RQL+
Sbjct: 361 MYPRHLVQPTNGMPSGNPLQTSANETPVLDQ--NASTKKSLGPAEHLQMQQPRQLNTPTP 418
Query: 350 LDGGSTKEVGSGNYAKPQGGPSQMPQKLNGFTKNQLHVLKAQILAFRRLKKGDGILPQEL 409
+ N + G +Q Q+ +GFTK QLHVLKAQILAFRRLKKG+G LP EL
Sbjct: 419 NLVAPSDTGPLSNSSLQSGQGTQQAQQRSGFTKQQLHVLKAQILAFRRLKKGEGSLPPEL 478
Query: 410 LEAISPPPLDLHVQQPIHSAGAQNQDKSMGNSVTEQPRQNEPKAKDSQPIVSFDGNS-SE 468
L+AISPPPL+L Q+ I A + QD+S + +Q R E K+SQ S +G S+
Sbjct: 479 LQAISPPPLELQTQRQISPAIGKVQDRSSDKTGEDQARSLEC-GKESQAAASSNGPIFSK 537
Query: 469 QETFVRDQKSTGAEVHMQAMLPVTKVS-----AGKEDQQSAGFSAKSDKKSEHVINRAPV 523
+E V D + H Q + K + A KE+QQ+ F KSD+ ++ + P
Sbjct: 538 EEDNVGDTEVALTTGHSQLFQNLGKEATSTDVATKEEQQTDVFPVKSDQGADSSTQKNPR 597
Query: 524 INDLALDKGKAVASQALVTDTAQINKPAQSSTVVGLPKDAGPAKKYYGPLFDFPFFTRKQ 583
+D DKGKAVAS D +Q P Q+++ PKD A+KYYGPLFDFPFFTRK
Sbjct: 598 -SDSTADKGKAVAS-----DGSQSKVPPQANSPQP-PKDTASARKYYGPLFDFPFFTRKL 650
Query: 584 DSFGSSMMANNNNNLSLAYDVKELLYEEGTEVFNKRRTENLKKIEGLLAVNLERKRIRPD 643
DS+GS+ AN NNNL+LAYD+K+L+ EEG E +K+RT++LKKI GLLA NLERKRIRPD
Sbjct: 651 DSYGSAT-ANANNNLTLAYDIKDLICEEGAEFLSKKRTDSLKKINGLLAKNLERKRIRPD 709
Query: 644 LVLKLQIEEKKLRLLDLQARLRGEIDQQQQEIMAMPDRPYRKFVKLCERQRVELARQVQT 703
LVL+LQIEEKKLRL DLQ+R+R E+D+QQQEIM+MPDRPYRKFV+LCERQR+E+ RQV
Sbjct: 710 LVLRLQIEEKKLRLSDLQSRVREEVDRQQQEIMSMPDRPYRKFVRLCERQRLEMNRQVLA 769
Query: 704 SQKALREKQLKSIFQWRKKLLEVHWAIRDARTARNRGVAKYHEKMLKEFSKNKDDDRNKR 763
+QKA+REKQLK+IFQWRKKLLE HWAIRDARTARNRGVAKYHEKML+EFSK KDD RNKR
Sbjct: 770 NQKAVREKQLKTIFQWRKKLLEAHWAIRDARTARNRGVAKYHEKMLREFSKRKDDGRNKR 829
Query: 764 MEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFLTQTEEYLQKLGSKITSAKNQQE 823
MEALKNNDV+RYREMLLEQQT++PGDAAERY VLS+FLTQTE+YL KLG KIT+ KNQQE
Sbjct: 830 MEALKNNDVERYREMLLEQQTNMPGDAAERYAVLSSFLTQTEDYLHKLGGKITATKNQQE 889
Query: 824 VEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIRNRFMEMNAPKDGSS 873
VEE+A AAA AARLQGLSEEEVRAAA CA EEV+IRNRF EMNAPK+ SS
Sbjct: 890 VEEAANAAAVAARLQGLSEEEVRAAATCAREEVVIRNRFTEMNAPKENSS 939
Score = 32.3 bits (72), Expect = 3.1
Identities = 44/178 (24%), Positives = 69/178 (38%), Gaps = 30/178 (16%)
Query: 96 SSSRNSSEHFSHGEKRVEQGQQLASDKK---------NEGKSSTQGLGIGHLMPGNNI-- 144
SSS +S + +++ +Q QQLAS ++ NE + Q G+ +M G N
Sbjct: 17 SSSSSSVQQQQQQQQQQQQQQQLASRQQQQQHRNSDTNENMFAYQPGGVQGMMGGGNFAS 76
Query: 145 RPVQALPTQQSIPIAMNNQIATSDQLRAMQAWAHERNIDLSQPANANFAAQLNLMQTRMV 204
P QQS + Q Q + ++N + Q A FA Q + +
Sbjct: 77 SPGSMQMPQQSRNFFESPQQQQQQQQQGSSTQEGQQNFNPMQQAYIQFAMQAQHQKAQ-- 134
Query: 205 QQSKESGAQSSSV-----------------PVSKQQATSPAVSSEGSAHANSSTDVSA 245
QQ++ SSSV P S+ QA+S S + A T+ S+
Sbjct: 135 QQARMGMVGSSSVGKDQDARMGMLNMQDLNPSSQPQASSSKPSGDQFARGERQTESSS 192
>At2g28290 putative SNF2 subfamily transcription regulator
Length = 1331
Score = 439 bits (1130), Expect = e-123
Identities = 390/1349 (28%), Positives = 614/1349 (44%), Gaps = 238/1349 (17%)
Query: 201 TRMVQQSKESGAQSSSVPVSKQQATSPAVSSEGSAHANSSTDVSALVGSVKARQTAPPSH 260
++ V S E A ++K A + GS A+ + + S ++ PS+
Sbjct: 98 SQAVGVSNEGKATLVENEMTKYDAFTSGRQLGGSNSASQTFYQGSGTQSNRSFDRESPSN 157
Query: 261 LGLPINAGVAGNSSDTAVQQFSLHGRDAQGSLKQLIVGVNGMPSMHPQQSSANKSLGADS 320
L N S+T Q RD + S K+ G S+ Q+ N +
Sbjct: 158 LDSTSGISQPHNRSETMNQ------RDVKSSGKRK----RGESSLSWDQNMDNSQIFDSH 207
Query: 321 SLNAKASSSRSDPEPAKMQYVRQLSQHASLDGGSTKEVG----------------SGNYA 364
++ + P +R L S D +T + G GN
Sbjct: 208 KIDDQTGEVSKIEMPGNSGDIRNLHVGLSSDAFTTPQCGWQSSEATAIRPAIHKEPGNNV 267
Query: 365 KPQGG-PSQMPQKLNGFTKNQLHVLKAQILAFRRLKKGDGILPQELLEAISPPPLDLHVQ 423
+G PS P F + QL L+AQ L F L+ +G++P++L HV+
Sbjct: 268 AGEGFLPSGSP-----FREQQLKQLRAQCLVFLALR--NGLVPKKL-----------HVE 309
Query: 424 QPIHSAGAQNQDKSMGNSVTEQPRQNEPKAKDSQPIVSFDGNSSEQETFVRDQKSTGAEV 483
+ N+ E+ F G + + G
Sbjct: 310 IALR------------NTFREED--------------GFRGELFDPKGRTHTSSDLGGIP 343
Query: 484 HMQAMLPVTKVSAGKEDQQSAGFSAKSDKKSEHVINRAPVINDLALDKGKAVASQALVTD 543
+ A+L T G+ D+ FS+K ++S + N + D K +AS+ + +
Sbjct: 344 DVSALLSRTDNPTGRLDEMD--FSSKETERSR--LGEKSFANTVFSDGQKLLASR-IPSS 398
Query: 544 TAQINKPAQSSTVV---GLPKDAGPAKKYYGPLF---DFPFFTRKQDSFGSSMMANNNNN 597
AQ S + GL K+ + + D + D F SS++ N +
Sbjct: 399 QAQTQVAVSHSQLTFSPGLTKNTPSEMVGWTGVIKTNDLSTSAVQLDEFHSSVLLTNLID 458
Query: 598 LSLAYDVKELLYEEGTEVF----------------NKR----RTENLKKIEGLLAVNLER 637
S D+ + EE + NKR R+ +LK+ + A+
Sbjct: 459 NSSNMDIWHIADEEEGNLQPSPKYTMSQKWIMGRQNKRLLVDRSWSLKQQKADQAIGSRF 518
Query: 638 KRIRPDLVL--------KLQIEEKKLRLLDLQARLRGEIDQQQQEIMAMPDRPYRKFVKL 689
++ + L K IE KKL+LL+LQ RLR E + +A + + K
Sbjct: 519 NELKESVSLSDDISAKTKSVIELKKLQLLNLQRRLRSEFVYNFFKPIATDVEHLKSYKKH 578
Query: 690 CERQRVELARQVQTSQKALREKQLKS--------IFQWRKKLLEVHWAIRDARTARNRGV 741
+R++ + + K R+++++ + ++KL ++ R+ NR
Sbjct: 579 KHGRRIKQLEKYEQKMKEERQRRIRERQKEFFGGLEVHKEKLEDLFKVRRERLKGFNRYA 638
Query: 742 AKYHEKMLKEFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFL 801
++H++ + + D + +++ LK NDV+ Y M+ + ++ + L
Sbjct: 639 KEFHKRKERLHREKIDKIQREKINLLKINDVEGYLRMVQDAKSDR----------VKQLL 688
Query: 802 TQTEEYLQKLGSKITSAKN-----QQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEV 856
+TE+YLQKLGSK+ AK + E +E+ + A +E+E A A
Sbjct: 689 KETEKYLQKLGSKLKEAKLLTSRFENEADETRTSNATDDETLIENEDESDQAKASYISAF 748
Query: 857 MIRNRFMEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLY 916
+ ++ + S KYY +AH++ E + QPS L G LREYQ+ GL+W++SLY
Sbjct: 749 FHLFLMVMIHTIQHYLESNEKYYLMAHSIKENINEQPSSLVGGKLREYQMNGLRWLVSLY 808
Query: 917 NNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFN------ 970
NN LNGILADEMGLGKTVQV++LI YLME K + GP L++VP++VL W+ N
Sbjct: 809 NNHLNGILADEMGLGKTVQVISLICYLMETKNDRGPFLVVVPSSVLPGWQSEINFWAPSI 868
Query: 971 --------PENC-------IDHAE*IAYMVTIGVM--------------HFLCWIEGSSV 1001
P+ I H + + T + H++ EG
Sbjct: 869 HKIVYCGTPDERRKLFKEQIVHQKFNVLLTTYEYLMNKHDRPKLSKIHWHYIIIDEG--- 925
Query: 1002 KIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKA 1061
R+K+ L DL Y RLLLTGTPLQN+L+ELW+LLN LLP +F++ +
Sbjct: 926 ------HRIKNASCKLNADLKHYVSSHRLLLTGTPLQNNLEELWALLNFLLPNIFNSSED 979
Query: 1062 FNDWFSKPFQKEDPNQNAEND-WLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKV 1120
F+ WF+KPFQ N E+ L E+ ++II+RLHQ+L PF+LRR +VE LP K+
Sbjct: 980 FSQWFNKPFQS-----NGESSALLSEEENLLIINRLHQVLRPFVLRRLKHKVENELPEKI 1034
Query: 1121 SIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKT 1180
++RC SA+Q + ++ + AK + ++N MELR
Sbjct: 1035 ERLIRCEASAYQKLLMKRVEDN---------------LGSIGNAKS-RAVHNSVMELRNI 1078
Query: 1181 CNHPLLNYPFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYL 1240
CNHP L+ + +V+ CGKL MLDR+L KL+ T HRVL FSTMT+LLD++E+YL
Sbjct: 1079 CNHPYLSQLHSEEHFLPPIVRLCGKLEMLDRMLPKLKATDHRVLFFSTMTRLLDVMEDYL 1138
Query: 1241 QWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDP 1300
+ Y R+DG T+ DR + I FN S FIFLLSIRA G G+NLQ+ADTV+++D
Sbjct: 1139 TLKGYKYLRLDGQTSGGDRGALIDGFNKSGSPFFIFLLSIRAGGVGVNLQAADTVILFDT 1198
Query: 1301 DPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKISSHQKEDEMRIGGTIDMEDELAGKD 1360
D NP+ + QA ARAHRIGQK++V V+ E V
Sbjct: 1199 DWNPQVDLQAQARAHRIGQKKDVLVLRFETV----------------------------- 1229
Query: 1361 RYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTTHEERRLTLETLLHDEERCQETVHD 1420
S+E +R++ ++K+ +A++ I AG FD T+ E+R+ LE+LL + ++ D
Sbjct: 1230 ---NSVEEQVRAS-AEHKLGVANQSITAGFFDNNTSAEDRKEYLESLLRESKK----EED 1281
Query: 1421 VPSLQE--VNRMIARNEEEVELFDQMDEE 1447
P L + +N +IAR E E+++F+ +D++
Sbjct: 1282 APVLDDDALNDLIARRESEIDIFESIDKQ 1310
>At5g19310 homeotic gene regulator - like protein
Length = 1041
Score = 439 bits (1128), Expect = e-122
Identities = 347/1053 (32%), Positives = 523/1053 (48%), Gaps = 203/1053 (19%)
Query: 587 GSSMMANNNNNLSLAYDV---KELLYEEGTEVFNKRRTE-------NLKKIEGLLAVNLE 636
G+S N + +S D + L YE G+ + + T L ++EGL + E
Sbjct: 60 GNSYTPNRGDLMSEFEDALLQQRLNYESGSRLAELKETRYKNRIHNRLSQLEGLPSNRGE 119
Query: 637 RKRIRPDLVLKLQIEEKKLRLLDLQARLRGEI-----------DQQQQ----EIMAMPDR 681
DL K +E L+L +LQ R+RGE+ D ++Q +M +P R
Sbjct: 120 ------DLQEKCLLELYGLKLQELQCRVRGEVSAEYWLRLNCADPERQLYDWGMMRLPRR 173
Query: 682 PYR---KFVKLCERQRVELARQVQTSQKALR-EKQLKSIFQW--RKKLLEVHWAIRDART 735
Y FV + Q R + +++ LR E++ K++ + RK EV A+R+ +
Sbjct: 174 MYGVGDSFVMEADDQ----FRNKRDAERLLRLEEEEKNLIETTQRKFFAEVLNAVREFQL 229
Query: 736 A----------RNRGVAKYHEKMLKEFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTS 785
RN GV +H K + ++ + R+ ALK++D + Y ++ E Q
Sbjct: 230 QIQASHRRCKQRNDGVQAWHGKQRQRATRAE----KLRIMALKSDDQEEYMKLAKEIQRQ 285
Query: 786 LPGDAAERYNVLSTFLTQTEEYLQKLGSKITSAKNQQEVEESAKAAAAAARLQGLSEEEV 845
+E +L S + +V+ + E+V
Sbjct: 286 KDAKLSENTKLLKG-----------------SESDLSDVD---------------APEDV 313
Query: 846 RAAAACAGEEVMIRNRFMEMNAPKDGSSSVSKYYNLA-HAVNEKVLRQPSMLRAGTLREY 904
A E++ + + N +G + +NLA H++ EKV +QPS+L+ G LR Y
Sbjct: 314 LPAQDI---EIIDSDNNDDSNDLLEGE----RQFNLAIHSIQEKVTKQPSLLQGGELRSY 366
Query: 905 QLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVN 964
QL GLQWM+SLYNN NGILADEMGLGKT+Q +ALIAYL+E K +GPHLI+ P AVL N
Sbjct: 367 QLEGLQWMVSLYNNDYNGILADEMGLGKTIQTIALIAYLLESKDLHGPHLILAPKAVLPN 426
Query: 965 WKCAF--------------NPENCIDHAE*IA----------YMVTIGVMHFLCWIEGSS 1000
W+ F + E + IA Y + + FL I+ +
Sbjct: 427 WENEFALWAPSISAFLYDGSKEKRTEIRARIAGGKFNVLITHYDLIMRDKAFLKKIDWNY 486
Query: 1001 VKIIFSTQRMKDRESVLARDLDR-YRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNK 1059
+ I+ R+K+ E LA+ L YR RRLLLTGTP+QN L+ELWSLLN LLP +F++
Sbjct: 487 M-IVDEGHRLKNHECALAKTLGTGYRIKRRLLLTGTPIQNSLQELWSLLNFLLPHIFNSI 545
Query: 1060 KAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPK 1119
F +WF+ PF + + L E++++II+RLH ++ PF+LRR+ EVE LP K
Sbjct: 546 HNFEEWFNTPFAE------CGSASLTDEEELLIINRLHHVIRPFLLRRKKSEVEKFLPGK 599
Query: 1120 VSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRK 1179
++L+C MSA+Q Y + G + L+ +S K+L N M+LRK
Sbjct: 600 TQVILKCDMSAWQKLYYKQVTDVGRVGLHSGNGKS-------------KSLQNLTMQLRK 646
Query: 1180 TCNHPLLNYPF-FSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEE 1238
CNHP L ++ K +V+ GK +LDR+L KL++ GHR+LLFS MT+L+D+LE
Sbjct: 647 CCNHPYLFVGADYNMCKKPEIVRASGKFELLDRLLPKLKKAGHRILLFSQMTRLIDLLEI 706
Query: 1239 YLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIY 1298
YL +Y R+DG+T + R + FN P+S F+FLLS RA G GLNLQ+ADT++I+
Sbjct: 707 YLSLNDYMYLRLDGSTKTDQRGILLKQFNEPDSPYFMFLLSTRAGGLGLNLQTADTIIIF 766
Query: 1299 DPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKISSHQKEDEMRIGGTIDMEDELAG 1358
D D NP+ ++QA RAHRIGQK+EV+V + +
Sbjct: 767 DSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVS---------------------------- 798
Query: 1359 KDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTTHEERRLTLETLLHDEERCQETV 1418
IGSIE +I +Q K+ + +VI AG F+ +T ++RR LE ++ +
Sbjct: 799 ----IGSIEEVILERAKQ-KMGIDAKVIQAGLFNTTSTAQDRREMLEEIM--SKGTSSLG 851
Query: 1419 HDVPSLQEVNRMIARNEEEVELFDQMDEE----EDWLEEMTRYDQVPDWIRAS------T 1468
DVPS +E+NR+ AR EEE +F+QMDEE E++ + +VP+W S T
Sbjct: 852 EDVPSEREINRLAARTEEEFWMFEQMDEERRKKENYKTRLMEEKEVPEWAYTSETQEDKT 911
Query: 1469 REVNAAIAASSKRPSKKNALSGGNVVL-----------DSTEIGSERRRGRPKGKKNPSY 1517
N + + KR K+ S L D++++ +R+R K + +
Sbjct: 912 NAKNHFGSLTGKRKRKEAVYSDSLSDLQWMKAMESEDEDASKVSQKRKRTDTKTRMSNGS 971
Query: 1518 K------ELEDSSEEISEDRNEDSAHDEGEIGE 1544
K E ++ EE E+R E+S + E E
Sbjct: 972 KAEAVLSESDEEKEEEEEERKEESGKESEEENE 1004
>At3g06010 putative transcriptional regulator
Length = 1132
Score = 437 bits (1123), Expect = e-122
Identities = 310/936 (33%), Positives = 481/936 (51%), Gaps = 147/936 (15%)
Query: 686 FVKLCERQRVELARQVQTSQKALREKQLKSIFQWRKKLLEVHWAIRDARTARNRGVAKYH 745
FV+ R E ++T+++ + L ++ +++ ++ A + R RN GV +H
Sbjct: 236 FVQRLSRLEEEEKNLIETAKRKFFAEVLNAVREFQLQIQ----ATQKRRRQRNDGVQAWH 291
Query: 746 EKMLKEFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFLTQTE 805
+ + ++ + R+ ALK++D + Y +++ E + ER L+T L +T
Sbjct: 292 GRQRQRATRAE----KLRLMALKSDDQEAYMKLVKESKN-------ER---LTTLLEETN 337
Query: 806 EYLQKLGSKIT---SAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIRNRF 862
+ L LG+ + AK + ++ + + + L E ++ ++ +
Sbjct: 338 KLLANLGAAVQRQKDAKLPEGIDLLKDSESDLSELDAPRSEPLQDLLPDQDIDITESDNN 397
Query: 863 MEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNG 922
+ N +G +Y + H++ EKV QPS+L G LR YQL GLQWM+SL+NN LNG
Sbjct: 398 DDSNDLLEGQR---QYNSAIHSIQEKVTEQPSLLEGGELRSYQLEGLQWMVSLFNNNLNG 454
Query: 923 ILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWK----------CAFNPE 972
ILADEMGLGKT+Q ++LIAYL+E KG GP+LI+ P AVL NW AF +
Sbjct: 455 ILADEMGLGKTIQTISLIAYLLENKGVPGPYLIVAPKAVLPNWVNEFATWVPSIAAFLYD 514
Query: 973 NCIDHAE*IAYMVT------IGVMHF-LCWIEGSSVK-------IIFSTQRMKDRESVLA 1018
++ + I + + + H+ L + + +K I+ R+K+ ES LA
Sbjct: 515 GRLEERKAIREKIAGEGKFNVLITHYDLIMRDKAFLKKIEWYYMIVDEGHRLKNHESALA 574
Query: 1019 RDL-DRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQ 1077
+ L YR RRLLLTGTP+QN L+ELWSLLN LLP +F++ + F +WF+ PF
Sbjct: 575 KTLLTGYRIKRRLLLTGTPIQNSLQELWSLLNFLLPHIFNSVQNFEEWFNAPFADRG--- 631
Query: 1078 NAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYD 1137
N L E++++IIHRLH ++ PF+LRR+ +EVE LP K ++L+C MSA+Q Y
Sbjct: 632 ---NVSLTDEEELLIIHRLHHVIRPFILRRKKDEVEKFLPGKTQVILKCDMSAWQKVYYK 688
Query: 1138 WIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPF-FSDLSK 1196
+ G + L +S K+L N M+LRK CNHP L ++ K
Sbjct: 689 QVTDMGRVGLQTGSGKS-------------KSLQNLTMQLRKCCNHPYLFVGGDYNMWKK 735
Query: 1197 DFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTAL 1256
+V+ GK +LDR+L KL++ GHR+LLFS MT+L+D+LE YL Y R+DGTT
Sbjct: 736 PEIVRASGKFELLDRLLPKLRKAGHRILLFSQMTRLIDVLEIYLTLNDYKYLRLDGTTKT 795
Query: 1257 EDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHR 1316
+ R + FN P+S F+FLLS RA G GLNLQ+ADTV+I+D D NP+ ++QA RAHR
Sbjct: 796 DQRGLLLKQFNEPDSPYFMFLLSTRAGGLGLNLQTADTVIIFDSDWNPQMDQQAEDRAHR 855
Query: 1317 IGQKREVKVIYMEAVVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQ 1376
IGQK+EV+V + +V GS+E +I +Q
Sbjct: 856 IGQKKEVRVFVLVSV--------------------------------GSVEEVILERAKQ 883
Query: 1377 YKIDMADEVINAGRFDQRTTHEERRLTLETLLHDEERCQETVHDVPSLQEVNRMIARNEE 1436
K+ + +VI AG F+ +T ++RR LE ++ T DVPS +E+NR+ AR+E+
Sbjct: 884 -KMGIDAKVIQAGLFNTTSTAQDRREMLEEIMRKGTSSLGT--DVPSEREINRLAARSED 940
Query: 1437 EVELFDQMDEE----EDWLEEMTRYDQVPDWIRASTREVNAAI-------AASSKRPSKK 1485
E +F++MDEE E++ + + +VP+W + + + + KR K+
Sbjct: 941 EFWMFERMDEERRRKENYRARLMQEQEVPEWAYTTQTQEEKLNNGKFHFGSVTGKRKRKE 1000
Query: 1486 -------------NALSGGNVVLDSTEIGSERRRGRPKGKKNPSYKELED-------SSE 1525
A+ G + + + R K + S K +E +SE
Sbjct: 1001 IVYSDTLSELQWLKAVESGEDLSKLSMRYNRREENASNTKTSTSKKVIESIQTVSDGTSE 1060
Query: 1526 EISEDRNE------------DSAHDEGEIGEFEDDG 1549
E E++ E D + +E E GE E+DG
Sbjct: 1061 EDEEEQEEERAKEMSGKQRVDKSEEEEEEGEEENDG 1096
>At3g06400 putative ATPase (ISW2-like)
Length = 1057
Score = 320 bits (819), Expect = 7e-87
Identities = 217/602 (36%), Positives = 313/602 (51%), Gaps = 92/602 (15%)
Query: 751 EFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFLTQTEEYLQK 810
E SK ++ R K M+ LK + +EML Q S+ D + +L Q E
Sbjct: 75 EISK-REKARLKEMQKLKKQKI---QEMLESQNASIDADMNNKGKGRLKYLLQQTELFAH 130
Query: 811 LGSKITSAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIRNRFMEMNAPKD 870
S+ + KA ++EEE EE + +D
Sbjct: 131 FAKSDGSSSQK-------KAKGRGRHASKITEEE-------EDEEYL--------KEEED 168
Query: 871 GSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGL 930
G L + N ++L QPS ++ G +R+YQL GL W++ LY N +NGILADEMGL
Sbjct: 169 G---------LTGSGNTRLLTQPSCIQ-GKMRDYQLAGLNWLIRLYENGINGILADEMGL 218
Query: 931 GKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNW--------------KCAFNPENCID 976
GKT+Q ++L+AYL E++G GPH+++ P + L NW K NPE
Sbjct: 219 GKTLQTISLLAYLHEYRGINGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLGNPEERRH 278
Query: 977 HAE*--IAYMVTIGVMHFLCWIEGSSVK--------IIFSTQRMKDRESVLARDLDRYRC 1026
E +A I V F I+ + II R+K+ S+L++ + +
Sbjct: 279 IREDLLVAGKFDICVTSFEMAIKEKTALRRFSWRYIIIDEAHRIKNENSLLSKTMRLFST 338
Query: 1027 HRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLET 1086
+ RLL+TGTPLQN+L ELW+LLN LLPE+F + + F++WF + END E
Sbjct: 339 NYRLLITGTPLQNNLHELWALLNFLLPEIFSSAETFDEWFQI---------SGENDQQE- 388
Query: 1087 EKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLR 1146
++ +LH++L PF+LRR +VE LPPK +L+ MS Q Y +
Sbjct: 389 -----VVQQLHKVLRPFLLRRLKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEA 443
Query: 1147 LNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNY---PFFSDLSKDFMVKCC 1203
+N E+ R L N M+LRK CNHP L P + D ++
Sbjct: 444 VNAGGERKR--------------LLNIAMQLRKCCNHPYLFQGAEPGPPYTTGDHLITNA 489
Query: 1204 GKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAI 1263
GK+ +LD++L KL+ RVL+FS MT+LLDILE+YL +R +Y RIDG T ++R+++I
Sbjct: 490 GKMVLLDKLLPKLKERDSRVLIFSQMTRLLDILEDYLMYRGYLYCRIDGNTGGDERDASI 549
Query: 1264 VDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREV 1323
+N P S+ F+FLLS RA G G+NL +AD V++YD D NP+ + QA RAHRIGQK+EV
Sbjct: 550 EAYNKPGSEKFVFLLSTRAGGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEV 609
Query: 1324 KV 1325
+V
Sbjct: 610 QV 611
>At5g18620 chromatin remodelling complex ATPase chain ISWI -like
protein
Length = 1069
Score = 313 bits (801), Expect = 9e-85
Identities = 190/465 (40%), Positives = 268/465 (56%), Gaps = 57/465 (12%)
Query: 888 KVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFK 947
++L QP+ ++ G LR+YQL GL W++ LY N +NGILADEMGLGKT+Q ++L+AYL E++
Sbjct: 182 RLLTQPACIQ-GKLRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLAYLHEYR 240
Query: 948 GNYGPHLIIVPNAVLVNW--------------KCAFNPENC--IDHAE*IAYMVTIGVMH 991
G GPH+++ P + L NW K NPE I +A I V
Sbjct: 241 GINGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLGNPEERRHIREELLVAGKFDICVTS 300
Query: 992 FLCWIEGSSVK--------IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKE 1043
F I+ + II R+K+ S+L++ + + + RLL+TGTPLQN+L E
Sbjct: 301 FEMAIKEKTTLRRFSWRYIIIDEAHRIKNENSLLSKTMRLFSTNYRLLITGTPLQNNLHE 360
Query: 1044 LWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPF 1103
LW+LLN LLPEVF + + F++WF + END E ++ +LH++L PF
Sbjct: 361 LWALLNFLLPEVFSSAETFDEWFQI---------SGENDQQE------VVQQLHKVLRPF 405
Query: 1104 MLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQ 1163
+LRR +VE LPPK +L+ MS Q Y + +N E+ R
Sbjct: 406 LLRRLKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEVVNGGGERKR-------- 457
Query: 1164 AKQYKTLNNRCMELRKTCNHPLLNY---PFFSDLSKDFMVKCCGKLWMLDRILIKLQRTG 1220
L N M+LRK CNHP L P + D +V GK+ +LD++L KL+
Sbjct: 458 ------LLNIAMQLRKCCNHPYLFQGAEPGPPYTTGDHLVTNAGKMVLLDKLLPKLKDRD 511
Query: 1221 HRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSI 1280
RVL+FS MT+LLDILE+YL +R Y RIDG T ++R+++I +N P S+ F+FLLS
Sbjct: 512 SRVLIFSQMTRLLDILEDYLMYRGYQYCRIDGNTGGDERDASIEAYNKPGSEKFVFLLST 571
Query: 1281 RAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
RA G G+NL +AD V++YD D NP+ + QA RAHRIGQK+EV+V
Sbjct: 572 RAGGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQV 616
>At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1)
Length = 764
Score = 272 bits (696), Expect = 1e-72
Identities = 209/656 (31%), Positives = 320/656 (47%), Gaps = 104/656 (15%)
Query: 730 IRDARTARNRGVAKYHEKMLKEFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPGD 789
+ + + V E++L +KN D A + + + RE E++ + G
Sbjct: 34 LNEEENCEEKSVTVVEEEIL--LAKNGDSSLISEAMAQEEEQLLKLRED--EEKANNAGS 89
Query: 790 AAE------RYNVLSTFLTQTEEYLQKLGSK--------ITSAKNQQEVEESAKAAAAAA 835
A ++ L LTQT+ Y + L K I S + E E++ + A
Sbjct: 90 AVAPNLNETQFTKLDELLTQTQLYSEFLLEKMEDITINGIESESQKAEPEKTGRGRKRKA 149
Query: 836 RLQGLSEEEVRAAAACAGEEVMIRNRFMEMNAPKDG---SSSVSKYYNLAHAVNEKVLRQ 892
Q + + RA AA M + +DG +S +++ + NE
Sbjct: 150 ASQYNNTKAKRAVAA------------MISRSKEDGETINSDLTEEETVIKLQNELC--- 194
Query: 893 PSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNY-- 950
+L G L+ YQL G++W++SL+ N LNGILAD+MGLGKT+Q + +++L KGN
Sbjct: 195 -PLLTGGQLKSYQLKGVKWLISLWQNGLNGILADQMGLGKTIQTIGFLSHL---KGNGLD 250
Query: 951 GPHLIIVPNAVLVNWK---CAFNP----------ENCIDHAE*IAYMVTIG--------- 988
GP+L+I P + L NW F P +N D T+G
Sbjct: 251 GPYLVIAPLSTLSNWFNEIARFTPSINAIIYHGDKNQRDELRRKHMPKTVGPKFPIVITS 310
Query: 989 ----------VMHFLCWIEGSSVKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQ 1038
++ W +I R+K+ + L R+L + +LLLTGTPLQ
Sbjct: 311 YEVAMNDAKRILRHYPW----KYVVIDEGHRLKNHKCKLLRELKHLKMDNKLLLTGTPLQ 366
Query: 1039 NDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQ 1098
N+L ELWSLLN +LP++F + F WF F +++ N+ + + E +++ ++ +LH
Sbjct: 367 NNLSELWSLLNFILPDIFTSHDEFESWFD--FSEKNKNEATKEE--EEKRRAQVVSKLHG 422
Query: 1099 ILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEK 1158
IL PF+LRR +VE SLP K I++ M+ Q + + + TL + E R +
Sbjct: 423 ILRPFILRRMKCDVELSLPRKKEIIMYATMTDHQKKFQEHLVNN-TLEAHLGENAIRGQG 481
Query: 1159 SPLYQAKQYKTLNNRCMELRKTCNHPLLN---------YPFFSDLSKDFMVKCCGKLWML 1209
++ K LNN ++LRK CNHP L YP ++ V CGK +L
Sbjct: 482 ---WKGK----LNNLVIQLRKNCNHPDLLQGQIDGSYLYPPVEEI-----VGQCGKFRLL 529
Query: 1210 DRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSP 1269
+R+L++L H+VL+FS TKLLDI++ Y + RIDG+ L++R I DF+
Sbjct: 530 ERLLVRLFANNHKVLIFSQWTKLLDIMDYYFSEKGFEVCRIDGSVKLDERRRQIKDFSDE 589
Query: 1270 NSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
S C IFLLS RA G G+NL +ADT ++YD D NP+ + QA+ R HRIGQ + V V
Sbjct: 590 KSSCSIFLLSTRAGGLGINLTAADTCILYDSDWNPQMDLQAMDRCHRIGQTKPVHV 645
>At2g13370 pseudogene
Length = 1738
Score = 271 bits (694), Expect = 2e-72
Identities = 191/592 (32%), Positives = 301/592 (50%), Gaps = 98/592 (16%)
Query: 888 KVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFK 947
K+ QP L GTLR+YQL GL ++++ + N N ILADEMGLGKTVQ ++++ +L +
Sbjct: 628 KLDEQPEWLIGGTLRDYQLEGLNFLVNSWLNDTNVILADEMGLGKTVQSVSMLGFLQNTQ 687
Query: 948 GNYGPHLIIVPNAVLVNWKCAF----------------------NPENCID--------H 977
GP L++VP + L NW F N N + +
Sbjct: 688 QIPGPFLVVVPLSTLANWAKEFRKWLPGMNIIVYVGTRASREVRNKTNDVHKVGRPIKFN 747
Query: 978 AE*IAYMVTI---GVMHFLCWIEGSSVKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTG 1034
A Y V + V+ + WI ++ R+K+ E+ L L + +LL+TG
Sbjct: 748 ALLTTYEVVLKDKAVLSKIKWI----YLMVDEAHRLKNSEAQLYTALLEFSTKNKLLITG 803
Query: 1035 TPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIH 1094
TPLQN ++ELW+LL+ L P F NK F + N + + E+E +
Sbjct: 804 TPLQNSVEELWALLHFLDPGKFKNKDEFVE-----------NYKNLSSFNESE-----LA 847
Query: 1095 RLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQS 1154
LH L P +LRR +++VE SLPPK+ +LR MS Q Y WI LN +
Sbjct: 848 NLHLELRPHILRRVIKDVEKSLPPKIERILRVEMSPLQKQYYKWILERNFHDLNKGVRGN 907
Query: 1155 RMEKSPLYQAKQYKTLNNRCMELRKTCNHPLL----NYPFFSDLSK----DFMVKCCGKL 1206
++ +L N +EL+K CNHP L ++ + D++ D ++ GKL
Sbjct: 908 QV------------SLLNIVVELKKCCNHPFLFESADHGYGGDINDNSKLDKIILSSGKL 955
Query: 1207 WMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDF 1266
+LD++L++L+ T HRVL+FS M ++LDIL EYL R ++R+DG+T E R+ A+ F
Sbjct: 956 VILDKLLVRLRETKHRVLIFSQMVRMLDILAEYLSLRGFQFQRLDGSTKAELRQQAMDHF 1015
Query: 1267 NSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV- 1325
N+P SD F FLLS RA G G+NL +ADTVVI+D D NP+N+ QA++RAHRIGQ+ V +
Sbjct: 1016 NAPASDDFCFLLSTRAGGLGINLATADTVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIY 1075
Query: 1326 --IYMEAVVDKISSHQKE---------DEMRIGGTIDMEDELAGKDRYIGSIESLIRSNI 1374
+ ++V ++I K ++ G ++ + G + + +++R
Sbjct: 1076 RFVTSKSVEEEILERAKRKMVLDHLVIQKLNAEGRLEKRETKKGSNFDKNELSAILRFG- 1134
Query: 1375 QQYKIDMADEVINAGRFDQRTTHEERRLTLETLLH-----DEERCQETVHDV 1421
A+E+ + D+ + + ++ +L +E+ ET H++
Sbjct: 1135 -------AEELFKEDKNDEESKKRLLSMDIDEILERAEQVEEKHTDETEHEL 1179
>At2g44980 putative transcription activator
Length = 861
Score = 221 bits (562), Expect = 5e-57
Identities = 178/571 (31%), Positives = 282/571 (49%), Gaps = 88/571 (15%)
Query: 900 TLREYQLVGLQWMLSLYNNKLNGILA-DEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVP 958
TL+ +Q+ G+ W++ Y +N +L D+MGLGKT+Q ++ ++YL +G GP L++ P
Sbjct: 50 TLKPHQVEGVSWLIQKYLLGVNVVLELDQMGLGKTLQAISFLSYLKFRQGLPGPFLVLCP 109
Query: 959 NAVLVNWKCAFNP---------------------ENCIDHAE*I-------AYMVTIGVM 990
+V W N ++ DH + Y + +
Sbjct: 110 LSVTDGWVSEINRFTPNLEVLRYVGDKYCRLDMRKSMYDHGHFLPFDVLLTTYDIALVDQ 169
Query: 991 HFLCWIEGSSVKIIFSTQRMKDRESVLARDL-DRYRCHRRLLLTGTPLQNDLKELWSLLN 1049
FL I II QR+K+ SVL L +++ RRLL+TGTP+QN+L ELW+L++
Sbjct: 170 DFLSQIPWQYA-IIDEAQRLKNPNSVLYNVLLEQFLIPRRLLITGTPIQNNLTELWALMH 228
Query: 1050 LLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRR- 1108
+P VF D F F++ + ND ET K L IL FMLRR
Sbjct: 229 FCMPLVFGTL----DQFLSAFKETGDGLDVSND-KETYKS------LKFILGAFMLRRTK 277
Query: 1109 ---VEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAK 1165
+E LPP + + + + Q IY T LR +E +E S
Sbjct: 278 SLLIESGNLVLPPLTELTVMVPLVSLQKKIY-----TSILR---KELPGLLELSS--GGS 327
Query: 1166 QYKTLNNRCMELRKTCNHPLLNYPFFSDL---SKDFMVKCCGKLWMLDRILIKLQRTGHR 1222
+ +L N ++LRK C+HP L +P + +V+ GKL +LD++L +L +GHR
Sbjct: 328 NHTSLQNIVIQLRKACSHPYL-FPGIEPEPFEEGEHLVQASGKLLVLDQLLKRLHDSGHR 386
Query: 1223 VLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNS----------PNSD 1272
VLLFS MT LDIL+++++ RR Y R+DG+ E+R +AI +F++ S+
Sbjct: 387 VLLFSQMTSTLDILQDFMELRRYSYERLDGSVRAEERFAAIKNFSAKTERGLDSEVDGSN 446
Query: 1273 CFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM--EA 1330
F+F++S RA G GLNL +ADTV+ Y+ D NP+ ++QA+ RAHRIGQ V I + E
Sbjct: 447 AFVFMISTRAGGVGLNLVAADTVIFYEQDWNPQVDKQALQRAHRIGQISHVLSINLVTEH 506
Query: 1331 VVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLI---------------RSNIQ 1375
V+++ + E ++++ + + D + K+ G + SL+ N++
Sbjct: 507 SVEEVILRRAERKLQLSHNV-VGDNMEEKEEDGGDLRSLVFGLQRFDPEEIHNEESDNLK 565
Query: 1376 QYKIDMADEVINAGRFDQRTTHEERRLTLET 1406
+I E + A R + EERR + +
Sbjct: 566 MVEISSLAEKVVAIRQNVEPDKEERRFEINS 596
>At5g44800 helicase-like protein
Length = 2228
Score = 217 bits (552), Expect = 7e-56
Identities = 163/534 (30%), Positives = 261/534 (48%), Gaps = 94/534 (17%)
Query: 892 QPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYG 951
QP LR G L +QL L W+ ++ N ILADEMGLGKTV A ++ L G
Sbjct: 666 QPQELRGGALFAHQLEALNWLRRCWHKSKNVILADEMGLGKTVSASAFLSSLYFEFGVAR 725
Query: 952 PHLIIVPNAVLVNWKCAFNPE----NCIDH------------------------AE*IAY 983
P L++VP + + NW F+ N +++ + +Y
Sbjct: 726 PCLVLVPLSTMPNWLSEFSLWAPLLNVVEYHGSAKGRAIIRDYEWHAKNSTGTTKKPTSY 785
Query: 984 MVTIGVMHFLCWIEGSS--------VKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGT 1035
+ + + + SS V ++ R+K+ ES L L+ + R+LLTGT
Sbjct: 786 KFNVLLTTYEMVLADSSHLRGVPWEVLVVDEGHRLKNSESKLFSLLNTFSFQHRVLLTGT 845
Query: 1036 PLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHR 1095
PLQN++ E+++LLN L P F + +F + F +D EK +
Sbjct: 846 PLQNNIGEMYNLLNFLQPSSFPSLSSFEERF--------------HDLTSAEK----VEE 887
Query: 1096 LHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSR 1155
L +++ P MLRR ++ ++PPK ++ +++ Q+ Y + + L
Sbjct: 888 LKKLVAPHMLRRLKKDAMQNIPPKTERMVPVELTSIQAEYYRAMLTKNYQILRN------ 941
Query: 1156 MEKSPLYQAKQYKTLNNRCMELRKTCNHPLL---NYPFFSDLS--KDFMVKCCGKLWMLD 1210
+ + +++ N M+LRK CNHP L P L D +K KL +L
Sbjct: 942 -----IGKGVAQQSMLNIVMQLRKVCNHPYLIPGTEPESGSLEFLHDMRIKASAKLTLLH 996
Query: 1211 RILIKLQRTGHRVLLFSTMTKLLDILEEYL--QWRRLVYRRIDGTTALEDRESAIVDFNS 1268
+L L + GHRVL+FS MTKLLDILE+YL ++ + R+DG+ A+ DR++AI FN
Sbjct: 997 SMLKVLHKEGHRVLIFSQMTKLLDILEDYLNIEFGPKTFERVDGSVAVADRQAAIARFNQ 1056
Query: 1269 PNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREV----- 1323
+ + F+FLLS RA G G+NL +ADTV+IYD D NP + QA+ RAHRIGQ + +
Sbjct: 1057 -DKNRFVFLLSTRACGLGINLATADTVIIYDSDFNPHADIQAMNRAHRIGQSKRLLVYRL 1115
Query: 1324 ----------------KVIYMEAVVDKISSHQKEDEMRIGGTIDMEDELAGKDR 1361
K++ + V+K S ++ +++ GT ++ ++ AG+++
Sbjct: 1116 VVRASVEERILQLAKKKLMLDQLFVNKSGSQKEFEDILRWGTEELFNDSAGENK 1169
>At2g25170 putative chromodomain-helicase-DNA-binding protein
Length = 1359
Score = 201 bits (510), Expect = 5e-51
Identities = 166/588 (28%), Positives = 280/588 (47%), Gaps = 89/588 (15%)
Query: 1003 IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAF 1062
I+ R+K+++S L L +Y + R+LLTGTPLQN+L EL+ L++ L F + + F
Sbjct: 420 IVDEGHRLKNKDSKLFSSLTQYSSNHRILLTGTPLQNNLDELFMLMHFLDAGKFGSLEEF 479
Query: 1063 NDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSI 1122
+ F +D NQ + I RLH++L P +LRR ++V +PPK +
Sbjct: 480 QEEF------KDINQEEQ------------ISRLHKMLAPHLLRRVKKDVMKDMPPKKEL 521
Query: 1123 VLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYK-----TLNNRCMEL 1177
+LR +S+ Q Y I + YQ K +LNN MEL
Sbjct: 522 ILRVDLSSLQKEYYKAIFTRN------------------YQVLTKKGGAQISLNNIMMEL 563
Query: 1178 RKTCNHPLLNY---PFFSDLSKDF--MVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKL 1232
RK C HP + P D ++ F +++ CGKL +LD++++KL+ GHRVL+++ +
Sbjct: 564 RKVCCHPYMLEGVEPVIHDANEAFKQLLESCGKLQLLDKMMVKLKEQGHRVLIYTQFQHM 623
Query: 1233 LDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSA 1292
LD+LE+Y ++ Y RIDG +R+ I FN+ NS+ F FLLS RA G G+NL +A
Sbjct: 624 LDLLEDYCTHKKWQYERIDGKVGGAERQIRIDRFNAKNSNKFCFLLSTRAGGLGINLATA 683
Query: 1293 DTVVIYDPDPNPKNEEQAVARAHRIGQKREV---KVIYMEAVVDKISSHQKEDEMRIGGT 1349
DTV+IYD D NP + QA+ARAHR+GQ +V ++I + +++ K+ +
Sbjct: 684 DTVIIYDSDWNPHADLQAMARAHRLGQTNKVMIYRLINRGTIEERMMQLTKKKMV----- 738
Query: 1350 IDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTTHEERRLTLETLLH 1409
+E + GK L NI Q ++ D++I G + + ++ + +
Sbjct: 739 --LEHLVVGK---------LKTQNINQEEL---DDIIRYGSKELFASEDDEAGKSGKIHY 784
Query: 1410 DEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEEEDWL---EEMTRYDQVPDWIRA 1466
D+ +++++ R+ E E DEEE+ ++ ++ + + A
Sbjct: 785 DD-------------AAIDKLLDRDLVEAEEVSVDDEEENGFLKAFKVANFEYIDENEAA 831
Query: 1467 STREVNAAIAASSKRPSKKNALSGGNVVLDSTEIGSERRRGRPKGKKNPSYKELEDSSE- 1525
+ A + S + A ++ D E+ + GK+ S K+L E
Sbjct: 832 ALEAQRVAAESKSSAGNSDRASYWEELLKDKFEL-HQAEELNALGKRKRSRKQLVSIEED 890
Query: 1526 EISEDRNEDSAHDEGEIGEFEDDGYSGAGI---AQPVDKDKLDDVTPS 1570
+++ + S DE E D +G G+ +P + D++ P+
Sbjct: 891 DLAGLEDVSSDGDESYEAESTDGEAAGQGVQTGRRPYRRKGRDNLEPT 938
Score = 63.2 bits (152), Expect = 2e-09
Identities = 34/71 (47%), Positives = 47/71 (65%), Gaps = 2/71 (2%)
Query: 899 GTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVP 958
G L YQL GL ++ ++ + + ILADEMGLGKT+Q +AL+A L F+ N PHL+I P
Sbjct: 271 GLLHPYQLEGLNFLRFSWSKQTHVILADEMGLGKTIQSIALLASL--FEENLIPHLVIAP 328
Query: 959 NAVLVNWKCAF 969
+ L NW+ F
Sbjct: 329 LSTLRNWEREF 339
>At3g57300 helicase-like protein
Length = 1507
Score = 194 bits (493), Expect = 5e-49
Identities = 167/580 (28%), Positives = 262/580 (44%), Gaps = 100/580 (17%)
Query: 684 RKFVKLCERQRVELARQVQTSQKALREKQLKSIFQWRKKLLEVHWAIRDARTARNRGVAK 743
++F C+R E+ +V S K R +++ R LL W D + A R K
Sbjct: 378 KRFADGCQR---EVRMKVGRSYKIPRTAPIRTRKISRDMLL--FWKRYDKQMAEER---K 429
Query: 744 YHEKMLKEFSKNKDDDRNKRMEALKNN----DVDRYREMLLEQQTSLPGDAAERYNVLST 799
EK E K + + R + + + N + Y + + S P +A +
Sbjct: 430 KQEKEAAEAFKREQEQRESKRQQQRLNFLIKQTELYSHFMQNKTDSNPSEALPIGDE--- 486
Query: 800 FLTQTEEYLQKLGSKITSAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIR 859
+E L + TSA EVE+ +A L E+ +RAA ++ I
Sbjct: 487 --NPIDEVLPE-----TSAAEPSEVEDPEEAE--------LKEKVLRAAQDAVSKQKQIT 531
Query: 860 NRF----------MEMNAPKD-----GSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREY 904
+ F EM P + GSS++ + V V Q L GTL+EY
Sbjct: 532 DAFDTEYMKLRQTSEMEGPLNDISVSGSSNIDLHNPSTMPVTSTV--QTPELFKGTLKEY 589
Query: 905 QLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVN 964
Q+ GLQW+++ Y LNGILADEMGLGKT+Q MA +A+L E K +GP L++ P +VL N
Sbjct: 590 QMKGLQWLVNCYEQGLNGILADEMGLGKTIQAMAFLAHLAEEKNIWGPFLVVAPASVLNN 649
Query: 965 W--------------------------KCAFNPENCID-----HAE*IAYMVTIGVMHFL 993
W + NP+ H +Y + + +
Sbjct: 650 WADEISRFCPDLKTLPYWGGLQERTILRKNINPKRMYRRDAGFHILITSYQLLVTDEKYF 709
Query: 994 CWIEGSSVKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLP 1053
++ + ++ Q +K S+ + L + C RLLLTGTP+QN++ ELW+LL+ ++P
Sbjct: 710 RRVKWQYM-VLDEAQAIKSSSSIRWKTLLSFNCRNRLLLTGTPIQNNMAELWALLHFIMP 768
Query: 1054 EVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVE 1113
+FDN FN+WFSK + +AE+ E + ++RLH IL+PFMLRR ++V
Sbjct: 769 MLFDNHDQFNEWFSKGIE-----NHAEHGGTLNEHQ---LNRLHAILKPFMLRRVKKDVV 820
Query: 1114 GSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNR 1173
L K + + C++S+ Q A Y IK+ +L + + + + K+ L N
Sbjct: 821 SELTTKTEVTVHCKLSSRQQAFYQAIKNKISLAELFDSNRGQ------FTDKKVLNLMNI 874
Query: 1174 CMELRKTCNHPLL-------NYPFFSDLSKDFMVKCCGKL 1206
++LRK CNHP L +Y +F S + G+L
Sbjct: 875 VIQLRKVCNHPELFERNEGSSYLYFGVTSNSLLPHPFGEL 914
Score = 126 bits (317), Expect = 1e-28
Identities = 95/296 (32%), Positives = 153/296 (51%), Gaps = 45/296 (15%)
Query: 1204 GKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAI 1263
GKL LD +L +L+ HRVLLF+ MTK+L+ILE+Y+ +R+ Y R+DG++ + DR +
Sbjct: 1206 GKLQTLDILLKRLRAGNHRVLLFAQMTKMLNILEDYMNYRKYKYLRLDGSSTIMDRRDMV 1265
Query: 1264 VDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREV 1323
DF SD F+FLLS RA G G+NL +ADTV+ Y+ D NP + QA+ RAHR+GQ ++V
Sbjct: 1266 RDFQH-RSDIFVFLLSTRAGGLGINLTAADTVIFYESDWNPTLDLQAMDRAHRLGQTKDV 1324
Query: 1324 ---KVIYMEAVVDKI--SSHQKEDEMRI---GGTIDMEDELAGKDRYIGSIESLIRSNIQ 1375
++I E V +KI + QK ++ GG + +D L D + SL+ + +
Sbjct: 1325 TVYRLICKETVEEKILHRASQKNTVQQLVMTGGHVQGDDFLGAAD-----VVSLLMDDAE 1379
Query: 1376 QYKIDMADEVINAGRFDQRTTHEERRLTLETLLHDEERCQETVHDVPSLQEVNRMIARNE 1435
A + +Q+ R L L+ +++ + D +
Sbjct: 1380 ------------AAQLEQKF----RELPLQVKDRQKKKTKRIRIDAEGDATL-------- 1415
Query: 1436 EEVELFDQMDEEEDWLEEMTRYDQVPDWIRASTREVNAAIAASSKRPSKKNALSGG 1491
EE+E D+ D ++ LEE P+ ++S ++ AA ++ P K + G
Sbjct: 1416 EELEDVDRQDNGQEPLEE-------PEKPKSSNKKRRAASNPKARAPQKAKEEANG 1464
>At2g02090 putative helicase
Length = 763
Score = 188 bits (477), Expect = 3e-47
Identities = 158/515 (30%), Positives = 236/515 (45%), Gaps = 109/515 (21%)
Query: 901 LREYQLVGLQWMLSLYNNKLNG-ILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPN 959
L+ YQLVG+ ++L LY + G ILADEMGLGKT+Q + + L + GPHL++ P
Sbjct: 213 LKPYQLVGVNFLLLLYKKGIEGAILADEMGLGKTIQAITYLTLLSRLNNDPGPHLVVCPA 272
Query: 960 AVLVNWK------CAFNPENCIDHAE*IAYMVTIGVMHFLCWIEGSSVKIIF------ST 1007
+VL NW+ C A AY + + +V ++ +
Sbjct: 273 SVLENWERELRKWCPSFTVLQYHGAARAAYSRELNSLSKAGKPPPFNVLLVCYSLFERHS 332
Query: 1008 QRMKDRESVLAR---------------DLDRYR----------CHRRLLLTGTPLQNDLK 1042
++ KD VL R D + YR ++RL+LTGTPLQNDL
Sbjct: 333 EQQKDDRKVLKRWRWSCVLMDEAHALKDKNSYRWKNLMSVARNANQRLMLTGTPLQNDLH 392
Query: 1043 ELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEP 1102
ELWSLL +LP++F + N K ED +I R+ IL P
Sbjct: 393 ELWSLLEFMLPDIFTTE---NVDLKKLLNAEDTE---------------LITRMKSILGP 434
Query: 1103 FMLRRRVEEVEGSLPPKVS-----IVLRCRMSAFQSAIYDWIKSTGT--LRLNPEEEQSR 1155
F+LRR +V L PK+ ++ R + A++ AI ++ ++ ++L+ + S
Sbjct: 435 FILRRLKSDVMQQLVPKIQRVEYVLMERKQEDAYKEAIEEYRAASQARLVKLSSKSLNSL 494
Query: 1156 MEKSPLYQAKQYKT----LNNRCMELRKTCN-----------HPLLNYPFFSDLSK---- 1196
+ P Q Y T + N + +R+ + HP+ + F L +
Sbjct: 495 AKALPKRQISNYFTQFRKIANHPLLIRRIYSDEDVIRIARKLHPIGAFGFECSLDRVIEE 554
Query: 1197 --------------------------DFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMT 1230
D V K L +L ++++GHRVL+FS T
Sbjct: 555 VKGFNDFRIHQLLFQYGVNDTKGTLSDKHVMLSAKCRTLAELLPSMKKSGHRVLIFSQWT 614
Query: 1231 KLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQ 1290
+LDILE L + YRR+DG+T + DR++ + FN+ S F LLS RA G+GLNL
Sbjct: 615 SMLDILEWTLDVIGVTYRRLDGSTQVTDRQTIVDTFNNDKS-IFACLLSTRAGGQGLNLT 673
Query: 1291 SADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
ADTV+I+D D NP+ + QA R HRIGQ + V +
Sbjct: 674 GADTVIIHDMDFNPQIDRQAEDRCHRIGQTKPVTI 708
>At3g19210 DNA repair protein, putative
Length = 688
Score = 177 bits (450), Expect = 5e-44
Identities = 147/494 (29%), Positives = 229/494 (45%), Gaps = 86/494 (17%)
Query: 901 LREYQLVGLQWMLSLYNN-----KLNG-ILADEMGLGKTVQVMALIAYLM--EFKGN--Y 950
LR +Q G+Q+M + +NG ILAD+MGLGKT+Q + L+ L+ F G
Sbjct: 76 LRPHQREGVQFMFDCVSGLHGSANINGCILADDMGLGKTLQSITLLYTLLCQGFDGTPMV 135
Query: 951 GPHLIIVPNAVLVNWKCAFNPENCIDHAE*IAYM------VTIGVMHF------------ 992
+I+ P +++ NW+ + D + IA V G+ F
Sbjct: 136 KKAIIVTPTSLVSNWEAEIK-KWVGDRIQLIALCESTRDDVLSGIDSFTRPRSALQVLII 194
Query: 993 -----------LCWIEGSSVKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDL 1041
C E + I R+K+ +++ R L C RR+LL+GTP+QNDL
Sbjct: 195 SYETFRMHSSKFCQSESCDLLICDEAHRLKNDQTLTNRALASLTCKRRVLLSGTPMQNDL 254
Query: 1042 KELWSLLNLLLPEVFDNKKAFNDWFSKPFQ-KEDPNQNAENDWLETEKKVIIIHRLHQIL 1100
+E ++++N P + F ++ P +P E L ++ + +++Q
Sbjct: 255 EEFFAMVNFTNPGSLGDAAHFRHYYEAPIICGREPTATEEEKNLAADRSAELSSKVNQ-- 312
Query: 1101 EPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSP 1160
F+LRR + LPPK+ V+ C+M+ QS +Y+ S+ L+ ++
Sbjct: 313 --FILRRTNALLSNHLPPKIIEVVCCKMTTLQSTLYNHFISSKNLK-----------RAL 359
Query: 1161 LYQAKQYKTLNNRCMELRKTCNHPLLNYP----------------------FFSDLSKDF 1198
AKQ K L L+K CNHP L Y FS S +
Sbjct: 360 ADNAKQTKVLAY-ITALKKLCNHPKLIYDTIKSGNPGTVGFENCLEFFPAEMFSGRSGAW 418
Query: 1199 M------VKCCGKLWMLDRILIKLQR-TGHRVLLFSTMTKLLDILEEYLQWRRLVYRRID 1251
V+ GK+ +L R+L L+R T R++L S T+ LD+ + + RR + R+D
Sbjct: 419 TGGDGAWVELSGKMHVLSRLLANLRRKTDDRIVLVSNYTQTLDLFAQLCRERRYPFLRLD 478
Query: 1252 GTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAV 1311
G+T + R+ + N P D F FLLS +A G GLNL A+ +V++DPD NP N++QA
Sbjct: 479 GSTTISKRQKLVNRLNDPTKDEFAFLLSSKAGGCGLNLIGANRLVLFDPDWNPANDKQAA 538
Query: 1312 ARAHRIGQKREVKV 1325
AR R GQK+ V V
Sbjct: 539 ARVWRDGQKKRVYV 552
>At3g12810 Snf2-related CBP activator protein, putative
Length = 1048
Score = 177 bits (449), Expect = 6e-44
Identities = 114/333 (34%), Positives = 167/333 (49%), Gaps = 69/333 (20%)
Query: 888 KVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFK 947
KV + L +LREYQ +GL W++++Y KLNGILADEMGLGKT+ +AL+A+L K
Sbjct: 108 KVRTKLPFLLKHSLREYQHIGLDWLVTMYEKKLNGILADEMGLGKTIMTIALLAHLACDK 167
Query: 948 GNYGPHLIIVPNAVLVNWKCAFNPENCIDHAE*IAYMVTIGVMHFLC-----------WI 996
G +GPHLI+VP +V++NW+ F + + ++ + W+
Sbjct: 168 GIWGPHLIVVPTSVMLNWETEF-----------LKWCPAFKILTYFGSAKERKLKRQGWM 216
Query: 997 EGSSVKIIFSTQRMKDRESVLAR------------------------DLDRYRCHRRLLL 1032
+ +S + +T R+ ++S + + L + RR+LL
Sbjct: 217 KLNSFHVCITTYRLVIQDSKMFKRKKWKYLILDEAHLIKNWKSQRWQTLLNFNSKRRILL 276
Query: 1033 TGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVII 1092
TGTPLQNDL ELWSL++ L+P VF + + F DWF P Q N +
Sbjct: 277 TGTPLQNDLMELWSLMHFLMPHVFQSHQEFKDWFCNPIAGMVEGQEKINK--------EV 328
Query: 1093 IHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIY-DWIKSTGTLRLNPEE 1151
I RLH +L PF+LRR +VE LP K V+ CR+S Q +Y D+I ST E
Sbjct: 329 IDRLHNVLRPFLLRRLKRDVEKQLPSKHEHVIFCRLSKRQRNLYEDFIAST--------E 380
Query: 1152 EQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHP 1184
Q+ + + + + M+LRK CNHP
Sbjct: 381 TQATLTSGSFF------GMISIIMQLRKVCNHP 407
Score = 115 bits (289), Expect = 2e-25
Identities = 59/123 (47%), Positives = 82/123 (65%), Gaps = 1/123 (0%)
Query: 1203 CGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESA 1262
CGKL L +L KL+ GHR L+F+ MTK+LD+LE ++ Y R+DG+T E+R++
Sbjct: 659 CGKLQELAMLLRKLKFGGHRALIFTQMTKMLDVLEAFINLYGYTYMRLDGSTPPEERQTL 718
Query: 1263 IVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKRE 1322
+ FN+ N F+F+LS R+ G G+NL ADTV+ YD D NP ++QA R HRIGQ RE
Sbjct: 719 MQRFNT-NPKIFLFILSTRSGGVGINLVGADTVIFYDSDWNPAMDQQAQDRCHRIGQTRE 777
Query: 1323 VKV 1325
V +
Sbjct: 778 VHI 780
>At3g54280 TATA box binding protein (TBP) associated factor (TAF)
-like protein
Length = 2049
Score = 168 bits (426), Expect = 3e-41
Identities = 167/597 (27%), Positives = 261/597 (42%), Gaps = 146/597 (24%)
Query: 901 LREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAY-LMEFKGNYG-----PHL 954
LR YQ G+ W+ L KL+GIL D+MGLGKT+Q A++A E +G+ P +
Sbjct: 1444 LRRYQQEGINWLGFLKRFKLHGILCDDMGLGKTLQASAIVASDAAERRGSTDELDVFPSI 1503
Query: 955 IIVPNAVLVNWKCAFNPENCID--------------------------HAE*IAYMVTIG 988
I+ P+ ++ +W AF E ID + +Y V
Sbjct: 1504 IVCPSTLVGHW--AFEIEKYIDLSLLSVLQYVGSAQDRVSLREQFNNHNVIITSYDVVRK 1561
Query: 989 VMHFLCWIEGSSVKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLL 1048
+ +L + I+ +K+ +S + + + + RL+L+GTP+QN++ ELWSL
Sbjct: 1562 DVDYLTQFSWNYC-ILDEGHIIKNAKSKITAAVKQLKAQHRLILSGTPIQNNIMELWSLF 1620
Query: 1049 NLLLPEVFDNKKAFNDWFSKP-FQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRR 1107
+ L+P ++ F + KP DP +A+ + E V+ + LH+ + PF+LRR
Sbjct: 1621 DFLMPGFLGTERQFQASYGKPLLAARDPKCSAK----DAEAGVLAMEALHKQVMPFLLRR 1676
Query: 1108 RVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGT-------LRLNPEEEQSRMEKSP 1160
EEV LP K+ C +S Q +Y+ + ++++ + + +P
Sbjct: 1677 TKEEVLSDLPEKIIQDRYCDLSPVQLKLYEQFSGSSAKQEISSIIKVDGSADSGNADVAP 1736
Query: 1161 ------LYQAKQYKTLNNRCMELRKTCNHPLL------NYPFFSDLSKDFMVKCCG---- 1204
++QA QY L K C+HPLL P SDL+ M+ C
Sbjct: 1737 TKASTHVFQALQY---------LLKLCSHPLLVLGDKVTEPVASDLAA--MINGCSDIIT 1785
Query: 1205 ---------KLWMLDRILIK-------------LQRTGHRVLLFSTMTKLLDILEEYL-- 1240
KL L IL + L HRVL+F+ LLDI+E+ L
Sbjct: 1786 ELHKVQHSPKLVALQEILEECGIGSDASSSDGTLSVGQHRVLIFAQHKALLDIIEKDLFQ 1845
Query: 1241 -QWRRLVYRRIDGTTALEDRESAIVDFNS-PNSDCFIFLLSIRAAGRGLNLQSADTVVIY 1298
+ + Y R+DG+ E R + FNS P D + LL+ G GLNL SADT+V
Sbjct: 1846 AHMKSVTYMRLDGSVVPEKRFEIVKAFNSDPTID--VLLLTTHVGGLGLNLTSADTLVFM 1903
Query: 1299 DPDPNPKNEE-----------QAVARAHRIGQKREVKVIYMEAVVDKISSHQKEDEMRIG 1347
+ D NP + QA+ RAHR+GQKR V V + MR
Sbjct: 1904 EHDWNPMRDHQFANIELNKLWQAMDRAHRLGQKRVVNVHRL--------------IMR-- 1947
Query: 1348 GTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTTHEERRLTL 1404
G++E + S +Q++K+ +A+ VINA +T + ++ L L
Sbjct: 1948 ----------------GTLEEKVMS-LQKFKVSVANTVINAENASMKTMNTDQLLDL 1987
>At1g03750 hypothetical protein
Length = 874
Score = 168 bits (425), Expect = 4e-41
Identities = 168/652 (25%), Positives = 294/652 (44%), Gaps = 129/652 (19%)
Query: 901 LREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYG--------- 951
L E+Q G+++M +LY N GIL D+MGLGKT+Q +A +A + G+ G
Sbjct: 151 LLEHQREGVKFMYNLYKNNHGGILGDDMGLGKTIQTIAFLAAVYGKDGDAGESCLLESDK 210
Query: 952 -PHLIIVPNAVLVNWKCAF---------------NPENCIDHAE*IAYMVTIGVMHFLCW 995
P LII P++++ NW+ F N + ++ + A V + V F +
Sbjct: 211 GPVLIICPSSIIHNWESEFSRWASFFKVSVYHGSNRDMILEKLK--ARGVEVLVTSFDTF 268
Query: 996 ------IEGSSVKIIFSTQ--RMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSL 1047
+ G + +I+ + + R+K+ +S L + +R+ LTGT +QN + EL++L
Sbjct: 269 RIQGPVLSGINWEIVIADEAHRLKNEKSKLYEACLEIKTKKRIGLTGTVMQNKISELFNL 328
Query: 1048 LNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRR 1107
+ P ++ F D++ +P + E +K+ L +L +MLRR
Sbjct: 329 FEWVAPGSLGTREHFRDFYDEPLKLGQRATAPERFVQIADKRK---QHLGSLLRKYMLRR 385
Query: 1108 RVEEVEGSLPP-KVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQ 1166
EE G L K V+ C+MS Q +Y + ++ ++ SPL Q++
Sbjct: 386 TKEETIGHLMMGKEDNVVFCQMSQLQRRVYQRMIQLPEIQCLVNKDNPCACGSPLKQSEC 445
Query: 1167 YKTL--------------NNRC------------MELRKTCNHPLLNYP----------- 1189
+ + ++ C M+L++ NH L P
Sbjct: 446 CRRIVPDGTIWSYLHRDNHDGCDSCPFCLVLPCLMKLQQISNHLELIKPNPKDEPEKQKK 505
Query: 1190 --------FFSDL--------SKDFM----VKCCGKLWMLDRILIKLQRTGHRVLLFSTM 1229
F +D+ SK FM VK CGK+ L++++ G ++LLFS
Sbjct: 506 DAEFVSTVFGTDIDLLGGISASKSFMDLSDVKHCGKMRALEKLMASWISKGDKILLFSYS 565
Query: 1230 TKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFN-SPNSDCFIFLLSIRAAGRGLN 1288
++LDILE++L + + R+DG+T R+S + DFN SP+ +FL+S +A G GLN
Sbjct: 566 VRMLDILEKFLIRKGYSFARLDGSTPTNLRQSLVDDFNASPSKQ--VFLISTKAGGLGLN 623
Query: 1289 LQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM-------EAVVDKISSHQKE 1341
L SA+ VVI+DP+ NP ++ QA R+ R GQKR V V + E V + Q+
Sbjct: 624 LVSANRVVIFDPNWNPSHDLQAQDRSFRYGQKRHVVVFRLLSAGSLEELVYTRQVYKQQL 683
Query: 1342 DEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKI-----DMADEVINAG----RFD 1392
+ + G ++ RY ++ + + I D++D++ + D
Sbjct: 684 SNIAVAGKME--------TRYFEGVQDCKEFQGELFGISNLFRDLSDKLFTSDIVELHRD 735
Query: 1393 QRTTHEERRLTLETLLHDEERCQETV------HDVPSLQEVNRMIARNEEEV 1438
++R LET + ++E+ +E + + P L+++ + A E++
Sbjct: 736 SNIDENKKRSLLETGVSEDEKEEEVMCSYKPEMEKPILKDLGIVYAHRNEDI 787
>At2g18760 putative SNF2/RAD54 family DNA repair and recombination
protein
Length = 1187
Score = 147 bits (370), Expect = 9e-35
Identities = 101/298 (33%), Positives = 153/298 (50%), Gaps = 20/298 (6%)
Query: 1029 RLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEK 1088
R+++TG P+QN L ELWSL + + P F FS P NA + T
Sbjct: 569 RIIMTGAPIQNKLTELWSLFDFVFPGKLGVLPVFEAEFSVPITVGG-YANASPLQVSTAY 627
Query: 1089 KVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLN 1148
+ ++ L ++ P++LRR +V L K VL C ++ Q + Y L +
Sbjct: 628 RCAVV--LRDLIMPYLLRRMKADVNAHLTKKTEHVLFCSLTVEQRSTY-----RAFLASS 680
Query: 1149 PEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSDLSKDF-MVKCCGKLW 1207
E+ ++ LY +RK CNHP L S + D+ + GK+
Sbjct: 681 EVEQIFDGNRNSLYGIDV----------MRKICNHPDLLEREHSHQNPDYGNPERSGKMK 730
Query: 1208 MLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFN 1267
++ +L ++ GHRVLLFS ++LDILE +L YRR+DG T ++ R + I +FN
Sbjct: 731 VVAEVLKVWKQQGHRVLLFSQTQQMLDILESFLVANEYSYRRMDGLTPVKQRMALIDEFN 790
Query: 1268 SPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
+ + D F+F+L+ + G G NL A+ V+I+DPD NP N+ QA RA RIGQK++V V
Sbjct: 791 N-SEDMFVFVLTTKVGGLGTNLTGANRVIIFDPDWNPSNDMQARERAWRIGQKKDVTV 847
Score = 61.2 bits (147), Expect = 6e-09
Identities = 42/159 (26%), Positives = 74/159 (46%), Gaps = 17/159 (10%)
Query: 809 QKLGSKITSAKNQQEVEESAKAAAAAARLQGLSE-EEVRAAAACAGEEVMIRNRFMEMNA 867
+K G K + E + + + + LQG + + ++C EE+ + F + +
Sbjct: 307 RKAGKKSKKTRPLPEKKWRKRISREDSSLQGSGDGRRILTTSSCEEEEL---DDFDDADD 363
Query: 868 PKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADE 927
+ S + N+ + K+ +YQ VG+QW+ L+ + GI+ DE
Sbjct: 364 NERSSVQLEGGLNIPECIFRKLF------------DYQRVGVQWLWELHCQRAGGIIGDE 411
Query: 928 MGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWK 966
MGLGKT+QV++ + L F Y P +II P +L W+
Sbjct: 412 MGLGKTIQVLSFLGSL-HFSKMYKPSIIICPVTLLRQWR 449
>At4g31900 putative protein
Length = 1067
Score = 145 bits (365), Expect = 3e-34
Identities = 121/445 (27%), Positives = 216/445 (48%), Gaps = 68/445 (15%)
Query: 1106 RRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAK 1165
R + + ++ +PPK ++LR MS+ Q +Y + + L + +
Sbjct: 326 RLKKDVLKDKVPPKKELILRVDMSSQQKEVYKAVITNNYQVLTKKRDAK----------- 374
Query: 1166 QYKTLNNRCMELRKTCNHPLLNYPF---FSDLSKDF--MVKCCGKLWMLDRILIKLQRTG 1220
++N M+LR+ C+HP L F F D ++ F +++ GKL +LD++++KL+ G
Sbjct: 375 ----ISNVLMKLRQVCSHPYLLPDFEPRFEDANEAFTKLLEASGKLQLLDKMMVKLKEQG 430
Query: 1221 HRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSI 1280
HRVL+++ L +LE+Y ++ Y RIDG + +R+ I FN+ NS+ F FLLS
Sbjct: 431 HRVLIYTQFQHTLYLLEDYFTFKNWNYERIDGKISGPERQVRIDRFNAENSNRFCFLLST 490
Query: 1281 RAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREV---KVIYMEAVVDKISS 1337
RA G G+NL +ADTV+IYD D NP + QA+AR HR+GQ +V ++I+ V +++
Sbjct: 491 RAGGIGINLATADTVIIYDSDWNPHADLQAMARVHRLGQTNKVMIYRLIHKGTVEERMME 550
Query: 1338 HQKEDEMRIGGTIDMEDELAGKDRYI-GSIESLIRSNIQQYKIDMADEVINAGRFDQRTT 1396
K + +E + GK ++ +I+ ++ + DE +G+
Sbjct: 551 ITKNKML-------LEHLVVGKQHLCQDELDDIIKYGSKELFSEENDEAGRSGK------ 597
Query: 1397 HEERRLTLETLLHDEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEEEDWLE--EM 1454
+H ++ E + D + V EV L D +EE D+L+ ++
Sbjct: 598 -----------IHYDDAAIEQLLDRNHVDAV---------EVSLDD--EEETDFLKNFKV 635
Query: 1455 TRYDQVPDWIRASTREV------NAAIAASSKRPSKKNALSGGNVVLDSTEIGSERRRGR 1508
++ V D A+ E N+++ + + K+ L V + E+ + +R R
Sbjct: 636 ASFEYVDDENEAAALEEAQAIENNSSVRNADRTSHWKDLLKDKYEVQQAEELSALGKRKR 695
Query: 1509 PKGKKNPSYKELEDSSEEISEDRNE 1533
GK+ ++ D EEIS++ +E
Sbjct: 696 -NGKQVMYAEDDLDGLEEISDEEDE 719
Score = 68.6 bits (166), Expect = 4e-11
Identities = 43/110 (39%), Positives = 63/110 (57%), Gaps = 7/110 (6%)
Query: 864 EMNAPKDGSSSVSKYYNLAHAVNEKVLRQ----PSMLRAGTLREYQLVGLQWMLSLYNNK 919
E+ KD +SS + + + N + +Q P L GTL YQL GL ++ ++ K
Sbjct: 185 EIQRFKDINSSSRRDKYVENERNREEFKQFDLTPEFL-TGTLHTYQLEGLNFLRYSWSKK 243
Query: 920 LNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAF 969
N ILADEMGLGKT+Q +A +A L F+ N PHL++ P + + NW+ F
Sbjct: 244 TNVILADEMGLGKTIQSIAFLASL--FEENLSPHLVVAPLSTIRNWEREF 291
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,487,775
Number of Sequences: 26719
Number of extensions: 2129885
Number of successful extensions: 7478
Number of sequences better than 10.0: 212
Number of HSP's better than 10.0 without gapping: 60
Number of HSP's successfully gapped in prelim test: 152
Number of HSP's that attempted gapping in prelim test: 6961
Number of HSP's gapped (non-prelim): 535
length of query: 2139
length of database: 11,318,596
effective HSP length: 115
effective length of query: 2024
effective length of database: 8,245,911
effective search space: 16689723864
effective search space used: 16689723864
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC139601.1


