

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139600.7 - phase: 0
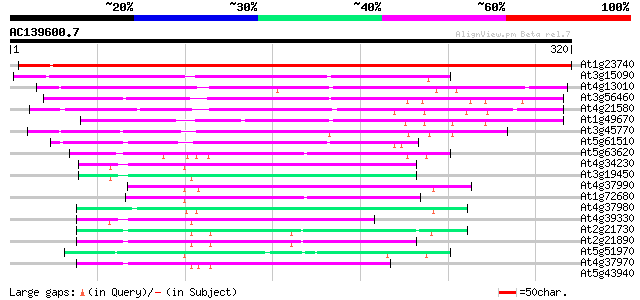
(320 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g23740 auxin-induced protein like 407 e-114
At3g15090 zinc-binding dehydrogenase, putative 138 3e-33
At4g13010 unknown protein 135 3e-32
At3g56460 quinone reductase-like protein 115 4e-26
At4g21580 putative NADPH quinone oxidoreductase 105 4e-23
At1g49670 ARP protein (At1g49670; F14J22.10) 91 8e-19
At3g45770 nuclear receptor binding factor-like protein 88 7e-18
At5g61510 quinone oxidoreductase - like protein 70 2e-12
At5g63620 alcohol dehydrogenase-like protein 67 1e-11
At4g34230 cinnamyl alcohol dehydrogenase like protein 66 3e-11
At3g19450 putative cinnamyl alcohol dehydrogenase 2 64 1e-10
At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2 62 3e-10
At1g72680 putative cinnamyl-alcohol dehydrogenase 62 3e-10
At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1 62 5e-10
At4g39330 cinnamyl-alcohol dehydrogenase CAD1 60 1e-09
At2g21730 putative cinnamyl-alcohol dehydrogenase 57 1e-08
At2g21890 putative cinnamyl-alcohol dehydrogenase 55 5e-08
At5g51970 sorbitol dehydrogenase-like protein 55 7e-08
At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa 55 7e-08
At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III (pir||S71... 39 0.003
>At1g23740 auxin-induced protein like
Length = 386
Score = 407 bits (1045), Expect = e-114
Identities = 203/315 (64%), Positives = 245/315 (77%), Gaps = 1/315 (0%)
Query: 6 SIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD 65
SIP KAWVYS YG ++ +LK +SN+ P KEDQVLIKVVAAALNPVD KR G FK
Sbjct: 73 SIPKEMKAWVYSDYGGVD-VLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQGKFKA 131
Query: 66 IDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEK 125
DSPLPTVPGYDVAGVVV VG VK K GDEVY +++E L PK G+L+EYT EEK
Sbjct: 132 TDSPLPTVPGYDVAGVVVKVGSAVKDLKEGDEVYANVSEKALEGPKQFGSLAEYTAVEEK 191
Query: 126 VLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVF 185
+LA KP N+ F +AA LPLAI TA +GL R EFS+GKS+LVL GAGGVGSLVIQLAKHV+
Sbjct: 192 LLALKPKNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLAKHVY 251
Query: 186 EASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEG 245
AS++AATAST KL+ +R LGADLAIDYTKEN E+L +K+DVV+DA+G ++AVK EG
Sbjct: 252 GASKVAATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEG 311
Query: 246 GKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYL 305
GKVV + TPP F++TS+G VL+KL PY+E+GKVKP++DPK PFPF + +AFSYL
Sbjct: 312 GKVVALTGAVTPPGFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYL 371
Query: 306 NTNRAIGKIVIHPIP 320
TN A GK+V++PIP
Sbjct: 372 ETNHATGKVVVYPIP 386
>At3g15090 zinc-binding dehydrogenase, putative
Length = 366
Score = 138 bits (348), Expect = 3e-33
Identities = 87/255 (34%), Positives = 137/255 (53%), Gaps = 14/255 (5%)
Query: 3 STPSIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGH 62
S S+ + +A + ++G E+ + NVP P+ ++VL+K A ++NP+D + G+
Sbjct: 24 SLRSVFTGCRAVILPRFGG-PEVFELRENVPVPNLNPNEVLVKAKAVSVNPLDCRIRAGY 82
Query: 63 FKDIDSP-LPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTV 121
+ + P LP + G DV+G V ++G VK KVG EV+G LH GT ++Y +
Sbjct: 83 GRSVFQPHLPIIVGRDVSGEVAAIGTSVKSLKVGQEVFG-----ALHPTALRGTYTDYGI 137
Query: 122 AEEKVLAHKPSNLSFVEAASLPLAIITAYQGLE-RVEFSSGKSLLVLGGAGGVGSLVIQL 180
E L KPS++S VEA+++P A +TA++ L+ + G+ LLV GG G VG IQL
Sbjct: 138 LSEDELTEKPSSISHVEASAIPFAALTAWRALKSNARITEGQRLLVFGGGGAVGFSAIQL 197
Query: 181 AKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYE-ELTEKFDVVYDAVGDSER-- 237
A V + A+ D + GA+ A+DYT E+ E + KFD V D +G E
Sbjct: 198 A--VASGCHVTASCVGQTKDRILAAGAEQAVDYTTEDIELAVKGKFDAVLDTIGGPETER 255
Query: 238 -AVKAANEGGKVVTI 251
+ +GG +T+
Sbjct: 256 IGINFLRKGGNYMTL 270
>At4g13010 unknown protein
Length = 329
Score = 135 bits (340), Expect = 3e-32
Identities = 103/330 (31%), Positives = 153/330 (46%), Gaps = 38/330 (11%)
Query: 16 YSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD-IDSPLPTVP 74
Y+ YG L+ VP P PK ++V +K+ A +LNPVD K G + + P +P
Sbjct: 11 YNSYGGGAAGLEH-VQVPVPTPKSNEVCLKLEATSLNPVDWKIQKGMIRPFLPRKFPCIP 69
Query: 75 GYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNL 134
DVAG VV VG VK FK GD+V ++ + G L+E+ VA EK+ +P +
Sbjct: 70 ATDVAGEVVEVGSGVKNFKAGDKVVAVLSHLGG------GGLAEFAVATEKLTVKRPQEV 123
Query: 135 SFVEAASLPLAIITAYQ------GLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEAS 188
EAA+LP+A +TA Q GL+ ++LV +GGVG +QLAK +
Sbjct: 124 GAAEAAALPVAGLTALQALTNPAGLKLDGTGKKANILVTAASGGVGHYAVQLAK--LANA 181
Query: 189 RIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAA------ 242
+ AT ++F++ LGAD +DY L YDAV + +
Sbjct: 182 HVTATCGARNIEFVKSLGADEVLDYKTPEGAALKSPSGKKYDAVVHCANGIPFSVFEPNL 241
Query: 243 NEGGKVVTILP--------------PGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILD 288
+E GKV+ I P +P LL LE + ++ GKVK ++D
Sbjct: 242 SENGKVIDITPGPNAMWTYAVKKITMSKKQLVPLLLIPKAENLEFMVNLVKEGKVKTVID 301
Query: 289 PKSPFPFYQTLEAFSYLNTNRAIGKIVIHP 318
K P + +A++ A GKI++ P
Sbjct: 302 SKHPLS--KAEDAWAKSIDGHATGKIIVEP 329
>At3g56460 quinone reductase-like protein
Length = 348
Score = 115 bits (287), Expect = 4e-26
Identities = 98/330 (29%), Positives = 154/330 (45%), Gaps = 44/330 (13%)
Query: 20 GNIEEILKFDSNVPTPHPKED-QVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDV 78
G+ E ++ P P D V ++V+A +LN + + LG +++ PLP +PG D
Sbjct: 18 GSPESPVEVSKTHPIPSLNSDTSVRVRVIATSLNYANYLQILGKYQE-KPPLPFIPGSDY 76
Query: 79 AGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVE 138
+G+V ++G V KF+VGD V + +G+ +++ VA++ L P V
Sbjct: 77 SGIVDAIGPAVTKFRVGDRVCSFAD---------LGSFAQFIVADQSRLFLVPERCDMVA 127
Query: 139 AASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTT 197
AA+LP+A T++ L R +SG+ LLVLG AGGVG +Q+ K V A IA T
Sbjct: 128 AAALPVAFGTSHVALVHRARLTSGQVLLVLGAAGGVGLAAVQIGK-VCGAIVIAVARGTE 186
Query: 198 KLDFLRKLGADLAIDYTKENYEELTEKF---------DVVYDAVGD--SERAVKAANEGG 246
K+ L+ +G D +D EN ++F DV+YD VG ++ ++K G
Sbjct: 187 KIQLLKSMGVDHVVDLGTENVISSVKEFIKTRKLKGVDVLYDPVGGKLTKESMKVLKWGA 246
Query: 247 KVVTI-LPPGTPPAIP--FLLTSDGAV-------LEKLQPYLENGKVKPILDPKS----- 291
+++ I G P IP L + V QP + +K +L S
Sbjct: 247 QILVIGFASGEIPVIPANIALVKNWTVHGLYWGSYRIHQPNVLEDSIKELLSWLSRGLIT 306
Query: 292 -----PFPFYQTLEAFSYLNTNRAIGKIVI 316
+ Q AF L +AIGK++I
Sbjct: 307 IHISHTYSLSQANLAFGDLKDRKAIGKVMI 336
>At4g21580 putative NADPH quinone oxidoreductase
Length = 325
Score = 105 bits (261), Expect = 4e-23
Identities = 99/338 (29%), Positives = 160/338 (47%), Gaps = 49/338 (14%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLP 71
KA V S+ G E + D V P K+D+VLI+V+A ALN D + LG + P
Sbjct: 2 KAIVISEPGKPEVLQLRD--VADPEVKDDEVLIRVLATALNRADTLQRLGLYNPPPGSSP 59
Query: 72 TVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKP 131
+ G + +G + SVG+ V ++KVGD+V ++ G +E + P
Sbjct: 60 YL-GLECSGTIESVGKGVSRWKVGDQVCALLSG---------GGYAEKVSVPAGQIFPIP 109
Query: 132 SNLSFVEAASLPLAIITAYQGLERV-EFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRI 190
+ +S +AA+ P T + + + S G+S L+ GG+ G+G+ IQ+AKH+ R+
Sbjct: 110 AGISLKDAAAFPEVACTVWSTVFMMGRLSVGESFLIHGGSSGIGTFAIQIAKHL--GVRV 167
Query: 191 AATA-STTKLDFLRKLGADLAIDYTKENY------EELTEKFDVVYDAVGDS--ERAVKA 241
TA S KL ++LGAD+ I+Y E++ E + DV+ D +G ++ + +
Sbjct: 168 FVTAGSDEKLAACKELGADVCINYKTEDFVAKVKAETDGKGVDVILDCIGAPYLQKNLDS 227
Query: 242 ANEGGKVVTILPPGTPPA-------IPFLLTSDGAVL----------------EKLQPYL 278
N G++ I G A +P LT GA L + + P +
Sbjct: 228 LNFDGRLCIIGLMGGANAEIKLSSLLPKRLTVLGAALRPRSPENKAVVVREVEKNVWPAI 287
Query: 279 ENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
E GKVKP++ P Q E S + ++ IGKI++
Sbjct: 288 EAGKVKPVI--YKYLPLSQAAEGHSLMESSNHIGKILL 323
>At1g49670 ARP protein (At1g49670; F14J22.10)
Length = 629
Score = 90.9 bits (224), Expect = 8e-19
Identities = 83/313 (26%), Positives = 138/313 (43%), Gaps = 51/313 (16%)
Query: 41 QVLIKVVAAALNPVDIKRALGHFKDIDSP-LPTVPGYDVAGVVVSVGEQVKKFKVGDEVY 99
QVL+K++ A +N D+ + G + SP LP G++ G++ +VGE VK +VG
Sbjct: 318 QVLLKIIYAGVNASDVNFSSGRYFTGGSPKLPFDAGFEGVGLIAAVGESVKNLEVG---- 373
Query: 100 GDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERV-EF 158
T T G SEY + K + P E ++ + +TA LE+ +
Sbjct: 374 ------TPAAVMTFGAYSEYMIVSSKHVLPVPR--PDPEVVAMLTSGLTALIALEKAGQM 425
Query: 159 SSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAAT-ASTTKLDFLRKLGADLAIDYTKEN 217
SG+++LV AGG G +QLAK +++ AT + K L++LG D IDY EN
Sbjct: 426 KSGETVLVTAAAGGTGQFAVQLAK--LSGNKVIATCGGSEKAKLLKELGVDRVIDYKSEN 483
Query: 218 YEELTEK-----FDVVYDAVGDS--ERAVKAANEGGKVVTI------------LPPGTPP 258
+ + +K +++Y++VG + + A G+++ I P P
Sbjct: 484 IKTVLKKEFPKGVNIIYESVGGQMFDMCLNALAVYGRLIVIGMISQYQGEKGWEPAKYPG 543
Query: 259 AIPFLLTSDGAV---------------LEKLQPYLENGKVKPILDPKSPFPFYQTLEAFS 303
+L V L+KL GK+K +D K +A
Sbjct: 544 LCEKILAKSQTVAGFFLVQYSQLWKQNLDKLFNLYALGKLKVGIDQKKFIGLNAVADAVE 603
Query: 304 YLNTNRAIGKIVI 316
YL++ ++ GK+V+
Sbjct: 604 YLHSGKSTGKVVV 616
>At3g45770 nuclear receptor binding factor-like protein
Length = 375
Score = 87.8 bits (216), Expect = 7e-18
Identities = 85/290 (29%), Positives = 134/290 (45%), Gaps = 26/290 (8%)
Query: 11 TKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPL 70
+KA VY ++G+ + + + N+P KE+ V +K++AA +NP DI R G + + P+
Sbjct: 45 SKAIVYEEHGSPDSVTRL-VNLPPVEVKENDVCVKMIAAPINPSDINRIEGVY-PVRPPV 102
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHK 130
P V GY+ G V +VG V F GD V + +P + GT Y V EE V
Sbjct: 103 PAVGGYEGVGEVYAVGSNVNGFSPGDWV--------IPSPPSSGTWQTYVVKEESVWHKI 154
Query: 131 PSNLSFVEAASLPLAIITAYQGLER-VEFSSGKSLLVLGGAGGVGSLVIQLA--KHVFEA 187
AA++ + +TA + LE V +SG S++ G VG VIQLA + +
Sbjct: 155 DKECPMEYAATITVNPLTALRMLEDFVNLNSGDSVVQNGATSIVGQCVIQLARLRGISTI 214
Query: 188 SRIAATASTTKL-DFLRKLGADLAIDYTKENYEELTEKFD------VVYDAVGDSERA-- 238
+ I A + + + L+ LGAD ++ N + + + ++ VG + +
Sbjct: 215 NLIRDRAGSDEAREQLKALGADEVFSESQLNVKNVKSLLGNLPEPALGFNCVGGNAASLV 274
Query: 239 VKAANEGGKVVTI----LPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVK 284
+K EGG +VT P T F+ LQ +L GKVK
Sbjct: 275 LKYLREGGTMVTYGGMSKKPITVSTTSFIFKDLALRGFWLQSWLSMGKVK 324
>At5g61510 quinone oxidoreductase - like protein
Length = 406
Score = 69.7 bits (169), Expect = 2e-12
Identities = 62/217 (28%), Positives = 106/217 (48%), Gaps = 19/217 (8%)
Query: 24 EILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVV 83
E+LK++ +V PKE ++ +K A LN +D+ G +K + +P PG + G VV
Sbjct: 96 EVLKWE-DVEVGEPKEGEIRVKNKAIGLNFIDVYFRKGVYKP--ASMPFTPGMEAVGEVV 152
Query: 84 SVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLP 143
+VG + +GD + + +G +E + + PS++ + AAS+
Sbjct: 153 AVGSGLTGRMIGD--------LVAYAGNPMGAYAEEQILPADKVVPVPSSIDPIVAASIM 204
Query: 144 LAIITAYQGLERV-EFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFL 202
L +TA L R + G ++LV AGGVGSL+ Q A + A+ I ++ K
Sbjct: 205 LKGMTAQFLLRRCFKVEPGHTILVHAAAGGVGSLLCQWA-NALGATVIGTVSTNEKAAQA 263
Query: 203 RKLGADLAIDYTKENY----EELT--EKFDVVYDAVG 233
++ G I Y E++ ++T + +VVYD+VG
Sbjct: 264 KEDGCHHVIMYKNEDFVSRVNDITSGKGVNVVYDSVG 300
>At5g63620 alcohol dehydrogenase-like protein
Length = 427
Score = 67.4 bits (163), Expect = 1e-11
Identities = 72/273 (26%), Positives = 114/273 (41%), Gaps = 60/273 (21%)
Query: 35 PHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVG-----EQV 89
P PK +++LIK A + D+ G +I P V G+++ G VV G + +
Sbjct: 76 PRPKSNEILIKTKACGVCHSDLHVMKG---EIPFASPCVIGHEITGEVVEHGPLTDHKII 132
Query: 90 KKFKVGDEVYG-----------------DINEI---------TLHNPKT----------- 112
+F +G V G D+ E TL++ +T
Sbjct: 133 NRFPIGSRVVGAFIMPCGTCSYCAKGHDDLCEDFFAYNRAKGTLYDGETRLFLRHDDSPV 192
Query: 113 ----IGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERV-EFSSGKSLLVL 167
+G ++EY V LA P +L + E+A L A+ TAY + E G S+ V+
Sbjct: 193 YMYSMGGMAEYCVTPAHGLAPLPESLPYSESAILGCAVFTAYGAMAHAAEIRPGDSIAVI 252
Query: 168 GGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKF-- 225
G GGVGS +Q+A+ + IA KL + LGA ++ KE+ E +
Sbjct: 253 G-IGGVGSSCLQIARAFGASDIIAVDVQDDKLQKAKTLGATHIVNAAKEDAVERIREITG 311
Query: 226 ----DVVYDAVGDSE---RAVKAANEGGKVVTI 251
DV +A+G + + + +GGK V I
Sbjct: 312 GMGVDVAVEALGKPQTFMQCTLSVKDGGKAVMI 344
>At4g34230 cinnamyl alcohol dehydrogenase like protein
Length = 357
Score = 65.9 bits (159), Expect = 3e-11
Identities = 57/223 (25%), Positives = 93/223 (41%), Gaps = 35/223 (15%)
Query: 40 DQVLIKVVAAALNPVDI---KRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGD 96
+ V I+++ + D+ K LG S P VPG++V G VV VG V KF VGD
Sbjct: 35 EDVNIRIICCGICHTDLHQTKNDLGM-----SNYPMVPGHEVVGEVVEVGSDVSKFTVGD 89
Query: 97 EV--------------------------YGDINEITLHNPKTIGTLSEYTVAEEKVLAHK 130
V N++ ++ T G ++ TV +K +
Sbjct: 90 IVGVGCLVGCCGGCSPCERDLEQYCPKKIWSYNDVYINGQPTQGGFAKATVVHQKFVVKI 149
Query: 131 PSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRI 190
P ++ +AA L A +T Y L + G GGVG + +++AK + +
Sbjct: 150 PEGMAVEQAAPLLCAGVTVYSPLSHFGLKQPGLRGGILGLGGVGHMGVKIAKAMGHHVTV 209
Query: 191 AATASTTKLDFLRKLGA-DLAIDYTKENYEELTEKFDVVYDAV 232
++++ + + L+ LGA D I + EL + D V D V
Sbjct: 210 ISSSNKKREEALQDLGADDYVIGSDQAKMSELADSLDYVIDTV 252
>At3g19450 putative cinnamyl alcohol dehydrogenase 2
Length = 365
Score = 63.5 bits (153), Expect = 1e-10
Identities = 55/223 (24%), Positives = 89/223 (39%), Gaps = 35/223 (15%)
Query: 40 DQVLIKVVAAALNPVDI---KRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGD 96
D V IKV+ + DI K LG S P VPG++V G V+ VG V KF VGD
Sbjct: 36 DDVYIKVICCGICHTDIHQIKNDLGM-----SNYPMVPGHEVVGEVLEVGSDVSKFTVGD 90
Query: 97 EVYGDI--------------------------NEITLHNPKTIGTLSEYTVAEEKVLAHK 130
V + N++ T G ++ + +K +
Sbjct: 91 VVGVGVVVGCCGSCKPCSSELEQYCNKRIWSYNDVYTDGKPTQGGFADTMIVNQKFVVKI 150
Query: 131 PSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRI 190
P ++ +AA L A +T Y L + + G GGVG + +++AK + +
Sbjct: 151 PEGMAVEQAAPLLCAGVTVYSPLSHFGLMASGLKGGILGLGGVGHMGVKIAKAMGHHVTV 210
Query: 191 AATASTTKLDFLRKLGA-DLAIDYTKENYEELTEKFDVVYDAV 232
+++ K + + LGA D + + L + D + D V
Sbjct: 211 ISSSDKKKEEAIEHLGADDYVVSSDPAEMQRLADSLDYIIDTV 253
>At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2
Length = 359
Score = 62.4 bits (150), Expect = 3e-10
Identities = 55/226 (24%), Positives = 92/226 (40%), Gaps = 30/226 (13%)
Query: 68 SPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV--------YGDINEIT------------- 106
S P VPG+++ GVV VG +V KFK G++V G + T
Sbjct: 60 STYPLVPGHEIVGVVTEVGAKVTKFKTGEKVGVGCLVSSCGSCDSCTEGMENYCPKSIQT 119
Query: 107 -----LHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSG 161
N T G S++ V EE + P NL AA L A IT Y ++
Sbjct: 120 YGFPYYDNTITYGGYSDHMVCEEGFVIRIPDNLPLDAAAPLLCAGITVYSPMKYHGLDKP 179
Query: 162 KSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGAD-LAIDYTKENYEE 220
+ + G GG+G + ++ AK + + +T+ + + + +LGAD + + ++
Sbjct: 180 GMHIGVVGLGGLGHVGVKFAKAMGTKVTVISTSEKKRDEAINRLGADAFLVSRDPKQIKD 239
Query: 221 LTEKFDVVYDAVGDSERAVK---AANEGGKVVTILPPGTPPAIPFL 263
D + D V + + GK+V + P P +P +
Sbjct: 240 AMGTMDGIIDTVSATHSLLPLLGLLKHKGKLVMVGAPEKPLELPVM 285
>At1g72680 putative cinnamyl-alcohol dehydrogenase
Length = 355
Score = 62.4 bits (150), Expect = 3e-10
Identities = 57/197 (28%), Positives = 84/197 (41%), Gaps = 30/197 (15%)
Query: 67 DSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV--------------------------YG 100
DS P VPG+++AG+V VG V++FKVGD V
Sbjct: 61 DSKYPLVPGHEIAGIVTKVGPNVQRFKVGDHVGVGTYVNSCRECEYCNEGQEVNCAKGVF 120
Query: 101 DINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSS 160
N I T G S + V E+ P + AA L A IT Y + R +
Sbjct: 121 TFNGIDHDGSVTKGGYSSHIVVHERYCYKIPVDYPLESAAPLLCAGITVYAPMMRHNMNQ 180
Query: 161 -GKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGAD-LAIDYTKENY 218
GKSL V+G GG+G + ++ K + + +T+ + K + L LGA+ I +
Sbjct: 181 PGKSLGVIG-LGGLGHMAVKFGKAFGLSVTVFSTSISKKEEALNLLGAENFVISSDHDQM 239
Query: 219 EELTEKFDVVYD-AVGD 234
+ L + D + D A GD
Sbjct: 240 KALEKSLDFLVDTASGD 256
>At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1
Length = 357
Score = 61.6 bits (148), Expect = 5e-10
Identities = 60/253 (23%), Positives = 100/253 (38%), Gaps = 32/253 (12%)
Query: 39 EDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV 98
E V KV+ + D+ A + PL VPG+++ GVV VG +VKKF GD+V
Sbjct: 33 EKDVRFKVLFCGICHTDLSMAKNEWGLTTYPL--VPGHEIVGVVTEVGAKVKKFNAGDKV 90
Query: 99 Y----------------GDINEI----------TLHNPKTIGTLSEYTVAEEKVLAHKPS 132
GD N + T G S++ V E + P
Sbjct: 91 GVGYMAGSCRSCDSCNDGDENYCPKMILTSGAKNFDDTMTHGGYSDHMVCAEDFIIRIPD 150
Query: 133 NLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAA 192
NL AA L A +T Y ++ + + G GG+G + ++ AK + + +
Sbjct: 151 NLPLDGAAPLLCAGVTVYSPMKYHGLDKPGMHIGVVGLGGLGHVAVKFAKAMGTKVTVIS 210
Query: 193 TASTTKLDFLRKLGAD-LAIDYTKENYEELTEKFDVVYDAVGDSERAVK---AANEGGKV 248
T+ + + + +LGAD + + ++ D + D V + + GK+
Sbjct: 211 TSERKRDEAVTRLGADAFLVSRDPKQMKDAMGTMDGIIDTVSATHPLLPLLGLLKNKGKL 270
Query: 249 VTILPPGTPPAIP 261
V + P P +P
Sbjct: 271 VMVGAPAEPLELP 283
>At4g39330 cinnamyl-alcohol dehydrogenase CAD1
Length = 360
Score = 60.5 bits (145), Expect = 1e-09
Identities = 50/199 (25%), Positives = 81/199 (40%), Gaps = 34/199 (17%)
Query: 39 EDQVLIKVVAAALNPVD---IKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVG 95
E+ V +K++ + D IK G+ S P VPG+++ G+ VG+ V KFK G
Sbjct: 37 ENDVTVKILFCGVCHTDLHTIKNDWGY-----SYYPVVPGHEIVGIATKVGKNVTKFKEG 91
Query: 96 DEVYGDI--------------------------NEITLHNPKTIGTLSEYTVAEEKVLAH 129
D V + N I K G SE V +++ +
Sbjct: 92 DRVGVGVISGSCQSCESCDQDLENYCPQMSFTYNAIGSDGTKNYGGYSENIVVDQRFVLR 151
Query: 130 KPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASR 189
P NL A L A IT Y ++ + L + G GG+G + +++ K
Sbjct: 152 FPENLPSDSGAPLLCAGITVYSPMKYYGMTEAGKHLGVAGLGGLGHVAVKIGKAFGLKVT 211
Query: 190 IAATASTTKLDFLRKLGAD 208
+ +++ST + + LGAD
Sbjct: 212 VISSSSTKAEEAINHLGAD 230
>At2g21730 putative cinnamyl-alcohol dehydrogenase
Length = 376
Score = 57.0 bits (136), Expect = 1e-08
Identities = 59/257 (22%), Positives = 105/257 (39%), Gaps = 38/257 (14%)
Query: 39 EDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV 98
E+ V +K++ + D+ H+ S P +PG+++ G+ VG+ V KFK GD V
Sbjct: 31 ENDVTVKILFCGVCHSDLHTIKNHWGF--SRYPIIPGHEIVGIATKVGKNVTKFKEGDRV 88
Query: 99 YGDI------------NEITLHNPKTI---------------GTLSEYTVAEEKVLAHKP 131
+ ++ + PK + G S+ V + + + P
Sbjct: 89 GVGVIIGSCQSCESCNQDLENYCPKVVFTYNSRSSDGTSRNQGGYSDVIVVDHRFVLSIP 148
Query: 132 SNLSFVEAASLPLAIITAYQGLERVEFS--SGKSLLVLGGAGGVGSLVIQLAKHVFEASR 189
L A L A IT Y ++ + SGK L V G GG+G + +++ K
Sbjct: 149 DGLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGV-NGLGGLGHIAVKIGKAFGLRVT 207
Query: 190 IAATASTTKLDFLRKLGAD-LAIDYTKENYEELTEKFDVVYDAVGDSERAV----KAANE 244
+ + +S + + + +LGAD + + +E D + D V +E A+
Sbjct: 208 VISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTMDFIIDTV-SAEHALLPLFSLLKV 266
Query: 245 GGKVVTILPPGTPPAIP 261
GK+V + P P +P
Sbjct: 267 NGKLVALGLPEKPLDLP 283
>At2g21890 putative cinnamyl-alcohol dehydrogenase
Length = 375
Score = 55.1 bits (131), Expect = 5e-08
Identities = 51/223 (22%), Positives = 92/223 (40%), Gaps = 32/223 (14%)
Query: 39 EDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV 98
E+ V +K++ + D+ H+ S P +PG+++ G+ VG+ V KFK GD V
Sbjct: 31 ENDVTVKILFCGVCHSDLHTIKNHWGF--SRYPIIPGHEIVGIATKVGKNVTKFKEGDRV 88
Query: 99 YGDI------------NEITLHNPKTI--------------GTLSEYTVAEEKVLAHKPS 132
+ ++ + PK + G S+ V + + + P
Sbjct: 89 GVGVIIGSCQSCESCNQDLENYCPKVVFTYNSRSSDGTRNQGGYSDVIVVDHRFVLSIPD 148
Query: 133 NLSFVEAASLPLAIITAYQGLERVEFS--SGKSLLVLGGAGGVGSLVIQLAKHVFEASRI 190
L A L A IT Y ++ + SGK L V G GG+G + +++ K +
Sbjct: 149 GLPSDSGAPLLCAGITVYSPMKYYGMTKESGKRLGV-NGLGGLGHIAVKIGKAFGLRVTV 207
Query: 191 AATASTTKLDFLRKLGAD-LAIDYTKENYEELTEKFDVVYDAV 232
+ +S + + + +LGAD + + +E D + D V
Sbjct: 208 ISRSSEKEREAIDRLGADSFLVTTDSQKMKEAVGTMDFIIDTV 250
>At5g51970 sorbitol dehydrogenase-like protein
Length = 364
Score = 54.7 bits (130), Expect = 7e-08
Identities = 65/254 (25%), Positives = 102/254 (39%), Gaps = 39/254 (15%)
Query: 32 VPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKK 91
+P+ P + +V +K V + V + + D P V G++ AG++ VGE+VK
Sbjct: 36 LPSVGPHDVRVRMKAVGICGSDVHYLKTM-RCADFVVKEPMVIGHECAGIIEEVGEEVKH 94
Query: 92 FKVGDEV------------------YGDINEITLH-NPKTIGTLSEYTVAEEKVLAHKPS 132
VGD V Y E+ P G+L+ V + P
Sbjct: 95 LVVGDRVALEPGISCWRCNLCREGRYNLCPEMKFFATPPVHGSLANQVVHPADLCFKLPE 154
Query: 133 NLSFVEAASL-PLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIA 191
N+S E A PL++ R E ++LV+ GAG +G LV LA F RI
Sbjct: 155 NVSLEEGAMCEPLSV--GVHACRRAEVGPETNVLVM-GAGPIG-LVTMLAARAFSVPRIV 210
Query: 192 -ATASTTKLDFLRKLGADLAIDYT----------KENYEELTEKFDVVYDAVGDSE---R 237
+L ++LGAD + T ++ + + DV +D G ++
Sbjct: 211 IVDVDENRLAVAKQLGADEIVQVTTNLEDVGSEVEQIQKAMGSNIDVTFDCAGFNKTMST 270
Query: 238 AVKAANEGGKVVTI 251
A+ A GGKV +
Sbjct: 271 ALAATRCGGKVCLV 284
>At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa
Length = 363
Score = 54.7 bits (130), Expect = 7e-08
Identities = 53/205 (25%), Positives = 89/205 (42%), Gaps = 28/205 (13%)
Query: 39 EDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEV 98
E++V +KV+ + D+ + S P VPG+++ G V +G +V KF +GD+V
Sbjct: 38 EEEVRVKVLYCGICHSDLHCLKNEWHS--SIYPLVPGHEIIGEVSEIGNKVSKFNLGDKV 95
Query: 99 -YGDI---------------NEIT--------LHNPKTI--GTLSEYTVAEEKVLAHKPS 132
G I N T +H+ TI G S++ V +E+ P
Sbjct: 96 GVGCIVDSCRTCESCREDQENYCTKAIATYNGVHHDGTINYGGYSDHIVVDERYAVKIPH 155
Query: 133 NLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAA 192
L V AA L A I+ Y ++ + + + G GG+G + ++ AK + +
Sbjct: 156 TLPLVSAAPLLCAGISMYSPMKYFGLTGPDKHVGIVGLGGLGHIGVRFAKAFGTKVTVVS 215
Query: 193 TASTTKLDFLRKLGADLAIDYTKEN 217
+ + D L LGAD + T E+
Sbjct: 216 STTGKSKDALDTLGADGFLVSTDED 240
>At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III
(pir||S71244)
Length = 379
Score = 39.3 bits (90), Expect = 0.003
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Query: 31 NVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVK 90
+V P+ +V IK++ AL D G KD + P + G++ AG+V SVGE V
Sbjct: 26 DVQVAPPQAGEVRIKILYTALCHTDAYTWSG--KDPEGLFPCILGHEAAGIVESVGEGVT 83
Query: 91 KFKVGDEV 98
+ + GD V
Sbjct: 84 EVQAGDHV 91
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.135 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,449,229
Number of Sequences: 26719
Number of extensions: 332724
Number of successful extensions: 767
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 695
Number of HSP's gapped (non-prelim): 56
length of query: 320
length of database: 11,318,596
effective HSP length: 99
effective length of query: 221
effective length of database: 8,673,415
effective search space: 1916824715
effective search space used: 1916824715
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139600.7


