

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.8 - phase: 0
(676 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
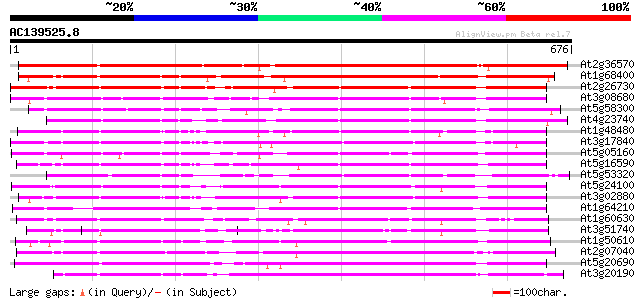
Score E
Sequences producing significant alignments: (bits) Value
At2g36570 putative receptor-like protein kinase 866 0.0
At1g68400 receptor kinase like protein 515 e-146
At2g26730 putative receptor-like protein kinase 474 e-134
At3g08680 putative protein kinase 461 e-130
At5g58300 receptor-like protein kinase 449 e-126
At4g23740 putative receptor kinase 427 e-120
At1g48480 protein kinase like protein 424 e-118
At3g17840 receptor kinase, putative 421 e-118
At5g05160 receptor-like protein kinase 410 e-114
At5g16590 receptor-like protein kinase 392 e-109
At5g53320 receptor protein kinase-like protein 392 e-109
At5g24100 receptor-like protein kinase 387 e-107
At3g02880 putative protein kinase 382 e-106
At1g64210 hypothetical protein 378 e-105
At1g60630 receptor kinase like protein 348 7e-96
At3g51740 unknown protein 330 1e-90
At1g50610 receptor-like protein kinase, putative 323 3e-88
At2g07040 putative leucine-rich repeat transmembrane protein kinase 322 3e-88
At5g20690 receptor protein kinase - like 319 3e-87
At3g20190 receptor kinase like protein 319 4e-87
>At2g36570 putative receptor-like protein kinase
Length = 672
Score = 866 bits (2238), Expect = 0.0
Identities = 455/675 (67%), Positives = 537/675 (79%), Gaps = 26/675 (3%)
Query: 11 LFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNN-RVTT 69
L L +++ L NDT ALTLFR QTDTHG L NWTG +AC++SW GV+C+P++ RVT
Sbjct: 10 LLLLLHLSITLAQNDTNALTLFRLQTDTHGNLAGNWTGSDACTSSWQGVSCSPSSHRVTE 69
Query: 70 LVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQI 129
L LPSL+LRGP+ +LSSL LRLLDLH+NRLNGTVS L+NC NL+L+YLAGND SG+I
Sbjct: 70 LSLPSLSLRGPLTSLSSLDQLRLLDLHDNRLNGTVSP--LTNCKNLRLVYLAGNDLSGEI 127
Query: 130 PPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTE 189
P EIS L ++RLDLSDNN+ G IP EI T +LT+R+QNN L+G IPD S M +L E
Sbjct: 128 PKEISFLKRMIRLDLSDNNIRGVIPREILGFTRVLTIRIQNNELTGRIPDFSQ-MKSLLE 186
Query: 190 LNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNP 249
LN++ NE +G V + ++ KFGD SFSGNEGLCGS P VC++T N P SS Q VPSNP
Sbjct: 187 LNVSFNELHGNVSDGVVKKFGDLSFSGNEGLCGSDPLPVCTIT-NDPESSNTDQIVPSNP 245
Query: 250 SSFPATSVIAR-PRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCAR----GRGVN 304
+S P + V R P H+G+ PG+I A++ CVA++V+ SF A CC R G
Sbjct: 246 TSIPHSPVSVREPEIHSHRGIKPGIIAAVIGG-CVAVIVLVSFGFAFCCGRLDRNGERSK 304
Query: 305 SNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRA 364
S S+ G G E K +S G GG+S D TS TD S+LVFF+RR FEL+DLL+A
Sbjct: 305 SGSVETGFVG-----GGEGKRRSSYGEGGES-DATSATDRSRLVFFERRKQFELDDLLKA 358
Query: 365 SAEMLGKGSLGTVYRAVLDDGST-VAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLR 423
SAEMLGKGSLGTVY+AVLDDGST VAVKRLKDANPC R EFEQYM++IG+LKH N+VKLR
Sbjct: 359 SAEMLGKGSLGTVYKAVLDDGSTTVAVKRLKDANPCPRKEFEQYMEIIGRLKHQNVVKLR 418
Query: 424 AYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYS 483
AYYYAKEEKLLVY+YL NGSLH+LLHGNRGPGRIPLDWTTRISL+LGAARGLA+IH EYS
Sbjct: 419 AYYYAKEEKLLVYEYLPNGSLHSLLHGNRGPGRIPLDWTTRISLMLGAARGLAKIHDEYS 478
Query: 484 AAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQA 543
+K+PHGN+KSSNVLLD+NGVA I+DFGLSLLLNPVHA ARLGGYRAPEQ+E KRLSQ+A
Sbjct: 479 ISKIPHGNIKSSNVLLDRNGVALIADFGLSLLLNPVHAIARLGGYRAPEQSEIKRLSQKA 538
Query: 544 DVYSFGVLLLEVLTGKAPSLQYPSPANRPRKV------EEEETVVDLPKWVRSVVREEWT 597
DVYSFGVLLLEVLTGKAPS+ +PSP +RPR EEEE VVDLPKWVRSVV+EEWT
Sbjct: 539 DVYSFGVLLLEVLTGKAPSI-FPSP-SRPRSAASVAVEEEEEAVVDLPKWVRSVVKEEWT 596
Query: 598 GEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDY 657
EVFD ELLRYKNIEEE+V+MLH+GLACVV QPEKRPTM +VVKM+E+IRVEQSP+ ED+
Sbjct: 597 AEVFDPELLRYKNIEEEMVAMLHIGLACVVPQPEKRPTMAEVVKMVEEIRVEQSPVGEDF 656
Query: 658 DESRNSLSPSIPTTE 672
DESRNS+SPS+ TT+
Sbjct: 657 DESRNSMSPSLATTD 671
>At1g68400 receptor kinase like protein
Length = 670
Score = 515 bits (1326), Expect = e-146
Identities = 309/670 (46%), Positives = 419/670 (62%), Gaps = 58/670 (8%)
Query: 11 LFLSIYIVPCL----THNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNR 66
L L I + CL + D++ L F+ D+ G+L + T C W GV+C NR
Sbjct: 13 LSLLILLQSCLLSSSSSTDSETLLNFKLTADSTGKLNSWNTTTNPCQ--WTGVSCN-RNR 69
Query: 67 VTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
VT LVL +NL G I +L+SLT LR+L L +N L+G + LSN T LKLL+L+ N FS
Sbjct: 70 VTRLVLEDINLTGSISSLTSLTSLRVLSLKHNNLSGPIPN--LSNLTALKLLFLSNNQFS 127
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G P I+SL L RLDLS NN +G IP +++ LT+LLTLRL++N SG IP+++ + +
Sbjct: 128 GNFPTSITSLTRLYRLDLSFNNFSGQIPPDLTDLTHLLTLRLESNRFSGQIPNIN--LSD 185
Query: 187 LTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSP---------- 236
L + N++ N F G++PN+ L++F + F+ N LCG+ P C+ + P
Sbjct: 186 LQDFNVSGNNFNGQIPNS-LSQFPESVFTQNPSLCGA-PLLKCTKLSSDPTKPGRPDEAK 243
Query: 237 --PSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVA 294
P ++P +TVPS+P TS+ +S + +S ++AI++ + L V S ++
Sbjct: 244 ASPLNKP-ETVPSSP-----TSIHGGDKSNNTSRISTISLIAIILGDFIILSFV-SLLLY 296
Query: 295 HCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSN----GGGGDSSDGTSGTDMSKLVFF 350
+C R VN K EK VY+SN +++ D K+VFF
Sbjct: 297 YCFWRQYAVNKKK-------HSKILEGEKIVYSSNPYPTSTQNNNNQNQQVGDKGKMVFF 349
Query: 351 DRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCA-RHEFEQYMD 409
+ FELEDLLRASAEMLGKG GT Y+AVL+DG+ VAVKRLKDA A + EFEQ M+
Sbjct: 350 EGTRRFELEDLLRASAEMLGKGGFGTAYKAVLEDGNEVAVKRLKDAVTVAGKKEFEQQME 409
Query: 410 VIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVL 469
V+G+L+H N+V L+AYY+A+EEKLLVYDY+ NGSL LLHGNRGPGR PLDWTTR+ +
Sbjct: 410 VLGRLRHTNLVSLKAYYFAREEKLLVYDYMPNGSLFWLLHGNRGPGRTPLDWTTRLKIAA 469
Query: 470 GAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYR 529
GAARGLA IH K+ HG++KS+NVLLD++G A +SDFGLS+ P A+ GYR
Sbjct: 470 GAARGLAFIHGSCKTLKLTHGDIKSTNVLLDRSGNARVSDFGLSIFA-PSQTVAKSNGYR 528
Query: 530 APEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVR 589
APE + ++ +Q++DVYSFGVLLLE+LTGK P++ + VDLP+WV+
Sbjct: 529 APELIDGRKHTQKSDVYSFGVLLLEILTGKCPNMV---------ETGHSGGAVDLPRWVQ 579
Query: 590 SVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR-- 647
SVVREEWT EVFD EL+RYK+IEEE+V +L + +AC + RP M VVK+IEDIR
Sbjct: 580 SVVREEWTAEVFDLELMRYKDIEEEMVGLLQIAMACTAVAADHRPKMGHVVKLIEDIRGG 639
Query: 648 -VEQSPLCED 656
E SP C D
Sbjct: 640 GSEASP-CND 648
>At2g26730 putative receptor-like protein kinase
Length = 658
Score = 474 bits (1220), Expect = e-134
Identities = 288/660 (43%), Positives = 401/660 (60%), Gaps = 58/660 (8%)
Query: 1 MNYVIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVT 60
+++V+ F + L V + + QAL F QQ +L N AC+ W GV
Sbjct: 4 ISWVLNSLFSILLLTQRVNSESTAEKQALLTFLQQIPHENRLQWN-ESDSACN--WVGVE 60
Query: 61 CTPN-NRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKL 117
C N + + +L LP L G P +L LT LR+L L +NRL+G + + SN T+L+
Sbjct: 61 CNSNQSSIHSLRLPGTGLVGQIPSGSLGRLTELRVLSLRSNRLSGQIPSDF-SNLTHLRS 119
Query: 118 LYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNI 177
LYL N+FSG+ P + LNNL+RLD+S NN G IP ++ LT+L L L NN SGN+
Sbjct: 120 LYLQHNEFSGEFPTSFTQLNNLIRLDISSNNFTGSIPFSVNNLTHLTGLFLGNNGFSGNL 179
Query: 178 PDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPP 237
P +S L + N++NN G +P++ L++F ESF+GN LCG P + C SP
Sbjct: 180 PSISL---GLVDFNVSNNNLNGSIPSS-LSRFSAESFTGNVDLCGG-PLKPCKSFFVSP- 233
Query: 238 SSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCC 297
S P PSN R S+ K LS IVAI+VA + L++ + ++ C
Sbjct: 234 SPSPSLINPSN-----------RLSSKKSK-LSKAAIVAIIVASALVALLLLALLLFLCL 281
Query: 298 ARGRGVN-SNSLMGSEAGKRK---------SYGSEKKVYNSNGGGGDSSDGTSGTDMSKL 347
+ RG N + + AG S E+ S+G GG+ T+ +KL
Sbjct: 282 RKRRGSNEARTKQPKPAGVATRNVDLPPGASSSKEEVTGTSSGMGGE-------TERNKL 334
Query: 348 VFFDRR-NGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQ 406
VF + F+LEDLLRASAE+LGKGS+GT Y+AVL++G+TV VKRLKD ++ EFE
Sbjct: 335 VFTEGGVYSFDLEDLLRASAEVLGKGSVGTSYKAVLEEGTTVVVKRLKDVM-ASKKEFET 393
Query: 407 YMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRIS 466
M+V+GK+KHPN++ LRAYYY+K+EKLLV+D++ GSL ALLHG+RG GR PLDW R+
Sbjct: 394 QMEVVGKIKHPNVIPLRAYYYSKDEKLLVFDFMPTGSLSALLHGSRGSGRTPLDWDNRMR 453
Query: 467 LVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLG 526
+ + AARGLA +H +AK+ HGN+K+SN+LL N C+SD+GL+ L + RL
Sbjct: 454 IAITAARGLAHLHV---SAKLVHGNIKASNILLHPNQDTCVSDYGLNQLFSNSSPPNRLA 510
Query: 527 GYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPK 586
GY APE E ++++ ++DVYSFGVLLLE+LTGK+P+ + E +DLP+
Sbjct: 511 GYHAPEVLETRKVTFKSDVYSFGVLLLELLTGKSPN-----------QASLGEEGIDLPR 559
Query: 587 WVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
WV SVVREEWT EVFD EL+RY NIEEE+V +L + +ACV P++RP M +V++MIED+
Sbjct: 560 WVLSVVREEWTAEVFDVELMRYHNIEEEMVQLLQIAMACVSTVPDQRPVMQEVLRMIEDV 619
>At3g08680 putative protein kinase
Length = 640
Score = 461 bits (1187), Expect = e-130
Identities = 285/659 (43%), Positives = 394/659 (59%), Gaps = 56/659 (8%)
Query: 1 MNYVIMFFFFLFLSIYIVPCLT---HNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWH 57
M +I F FL ++ ++ CL+ +D QAL F +L NW ASW
Sbjct: 1 MMKIIAAFLFLLVTTFVSRCLSADIESDKQALLEFASLVPHSRKL--NWNSTIPICASWT 58
Query: 58 GVTCTPNN-RVTTLVLPSLNLRGPID--ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTN 114
G+TC+ NN RVT L LP L GP+ L LR++ L +N L G + + +LS
Sbjct: 59 GITCSKNNARVTALRLPGSGLYGPLPEKTFEKLDALRIISLRSNHLQGNIPSVILS-LPF 117
Query: 115 LKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALS 174
++ LY N+FSG IPP +S + L+ LDLS N+L+G+IP + LT L L LQNN+LS
Sbjct: 118 IRSLYFHENNFSGTIPPVLS--HRLVNLDLSANSLSGNIPTSLQNLTQLTDLSLQNNSLS 175
Query: 175 GNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTEN 234
G IP+L P L LN++ N G VP+++ F SF GN LCG+ P C
Sbjct: 176 GPIPNLP---PRLKYLNLSFNNLNGSVPSSV-KSFPASSFQGNSLLCGA-PLTPCPENTT 230
Query: 235 SPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVA 294
+P S P+ P+ P T+ I R ++ K LS G IV I V V L ++ + ++
Sbjct: 231 APSPS------PTTPTEGPGTTNIGRGTAK--KVLSTGAIVGIAVGGSVLLFIILA-IIT 281
Query: 295 HCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN 354
CCA+ R + G+ + + K S+ + G + +KLVFF+ +
Sbjct: 282 LCCAKKR----------DGGQDSTAVPKAKPGRSDNKAEEFGSGVQEAEKNKLVFFEGSS 331
Query: 355 -GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGK 413
F+LEDLLRASAE+LGKGS GT Y+A+L++G+TV VKRLK+ + EFEQ M+ +G+
Sbjct: 332 YNFDLEDLLRASAEVLGKGSYGTTYKAILEEGTTVVVKRLKEV-AAGKREFEQQMEAVGR 390
Query: 414 LK-HPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAA 472
+ H N+ LRAYY++K+EKLLVYDY G+ LLHGN GR LDW TR+ + L AA
Sbjct: 391 ISPHVNVAPLRAYYFSKDEKLLVYDYYQGGNFSMLLHGNNEGGRAALDWETRLRICLEAA 450
Query: 473 RGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHAT---ARLGGYR 529
RG++ IH+ S AK+ HGN+KS NVLL + C+SDFG++ L++ H T +R GYR
Sbjct: 451 RGISHIHSA-SGAKLLHGNIKSPNVLLTQELHVCVSDFGIAPLMS--HHTLIPSRSLGYR 507
Query: 530 APEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVR 589
APE E ++ +Q++DVYSFGVLLLE+LTGKA K E VVDLPKWV+
Sbjct: 508 APEAIETRKHTQKSDVYSFGVLLLEMLTGKAAG-----------KTTGHEEVVDLPKWVQ 556
Query: 590 SVVREEWTGEVFDQELLRYK-NIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
SVVREEWTGEVFD EL++ + N+EEE+V ML + +ACV + P+ RP+M +VV M+E+IR
Sbjct: 557 SVVREEWTGEVFDVELIKQQHNVEEEMVQMLQIAMACVSKHPDSRPSMEEVVNMMEEIR 615
>At5g58300 receptor-like protein kinase
Length = 654
Score = 449 bits (1154), Expect = e-126
Identities = 279/653 (42%), Positives = 390/653 (58%), Gaps = 60/653 (9%)
Query: 23 HNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNR-VTTLVLPSLNLRGPI 81
++D QAL F +L NW SW GVTCT + V L LP + L GPI
Sbjct: 46 NSDRQALLAFAASVPHLRRL--NWNSTNHICKSWVGVTCTSDGTSVHALRLPGIGLLGPI 103
Query: 82 --DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNL 139
+ L L LR+L L +N L+G + + S +L +YL N+FSG++P +S N+
Sbjct: 104 PPNTLGKLESLRILSLRSNLLSGNLPPDIHS-LPSLDYIYLQHNNFSGEVPSFVSRQLNI 162
Query: 140 LRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYG 199
L DLS N+ G IP L L L LQNN LSG +P+L ++ +L LN++NN G
Sbjct: 163 L--DLSFNSFTGKIPATFQNLKQLTGLSLQNNKLSGPVPNLDTV--SLRRLNLSNNHLNG 218
Query: 200 KVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIA 259
+P+ L F SFSGN LCG P Q C+ T + PPS P + P P FP
Sbjct: 219 SIPSA-LGGFPSSSFSGNTLLCGL-PLQPCA-TSSPPPSLTPHISTPPLPP-FP------ 268
Query: 260 RPRSQHHKGLSPGVIVAIVVAICV---ALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKR 316
H +G + V+ ++ I ALL++ + ++ CC + + +S++ +
Sbjct: 269 -----HKEGSKRKLHVSTIIPIAAGGAALLLLITVIILCCCIKKKDKREDSIVKVKTLTE 323
Query: 317 KSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLG 375
K+ K+ + S G + +KLVFF+ + F+LEDLLRASAE+LGKGS G
Sbjct: 324 KA----KQEFGS---------GVQEPEKNKLVFFNGCSYNFDLEDLLRASAEVLGKGSYG 370
Query: 376 TVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKL-KHPNIVKLRAYYYAKEEKLL 434
T Y+AVL++ +TV VKRLK+ + EFEQ M++I ++ HP++V LRAYYY+K+EKL+
Sbjct: 371 TAYKAVLEESTTVVVKRLKEV-AAGKREFEQQMEIISRVGNHPSVVPLRAYYYSKDEKLM 429
Query: 435 VYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKS 494
V DY G+L +LLHGNRG + PLDW +R+ + L AA+G+A +H K HGN+KS
Sbjct: 430 VCDYYPAGNLSSLLHGNRGSEKTPLDWDSRVKITLSAAKGIAHLHAA-GGPKFSHGNIKS 488
Query: 495 SNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLE 554
SNV++ + ACISDFGL+ L+ A R GYRAPE E ++ + ++DVYSFGVL+LE
Sbjct: 489 SNVIMKQESDACISDFGLTPLMAVPIAPMRGAGYRAPEVMETRKHTHKSDVYSFGVLILE 548
Query: 555 VLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEE 614
+LTGK+P +Q PS + +VDLP+WV+SVVREEWT EVFD EL+R++NIEEE
Sbjct: 549 MLTGKSP-VQSPS----------RDDMVDLPRWVQSVVREEWTSEVFDIELMRFQNIEEE 597
Query: 615 LVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQS----PLCEDYDESRNS 663
+V ML + +ACV Q PE RPTM DVV+MIE+IRV S P +D + ++S
Sbjct: 598 MVQMLQIAMACVAQVPEVRPTMDDVVRMIEEIRVSDSETTRPSSDDNSKPKDS 650
>At4g23740 putative receptor kinase
Length = 638
Score = 427 bits (1098), Expect = e-120
Identities = 251/637 (39%), Positives = 373/637 (58%), Gaps = 55/637 (8%)
Query: 45 NWTGPEACSASWHGVTCTPN-NRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLN 101
NW W GVTC + +R+ + LP + L G P + +S L+ LR+L L +N ++
Sbjct: 47 NWNETSQVCNIWTGVTCNQDGSRIIAVRLPGVGLNGQIPPNTISRLSALRVLSLRSNLIS 106
Query: 102 GTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLT 161
G + +L LYL N+ SG +P + S NL ++LS+N G IP+ +SRL
Sbjct: 107 GEFPKDFVE-LKDLAFLYLQDNNLSGPLPLDFSVWKNLTSVNLSNNGFNGTIPSSLSRLK 165
Query: 162 NLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNN-EFYGKVPNTMLNKFGDESFSGNEGL 220
+ +L L NN LSG+IPDLS ++ +L ++++NN + G +P+ L +F S++G + +
Sbjct: 166 RIQSLNLANNTLSGDIPDLS-VLSSLQHIDLSNNYDLAGPIPD-WLRRFPFSSYTGIDII 223
Query: 221 CGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVA 280
PP P PS +P GLS V + IV+A
Sbjct: 224 ---------------PPGGNYTLVTPPPPSE----QTHQKPSKARFLGLSETVFLLIVIA 264
Query: 281 ICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTS 340
+ + ++ +FV+ C R + + ++ S+ K+ G +
Sbjct: 265 VSIVVITALAFVLTVCYVRRKLRRGDGVI-----------SDNKLQKKGGMSPEKFVSRM 313
Query: 341 GTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPC 399
++L FF+ N F+LEDLLRASAE+LGKG+ GT Y+AVL+D ++VAVKRLKD
Sbjct: 314 EDVNNRLSFFEGCNYSFDLEDLLRASAEVLGKGTFGTTYKAVLEDATSVAVKRLKDV-AA 372
Query: 400 ARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPL 459
+ +FEQ M++IG +KH N+V+L+AYYY+K+EKL+VYDY S GS+ +LLHGNRG RIPL
Sbjct: 373 GKRDFEQQMEIIGGIKHENVVELKAYYYSKDEKLMVYDYFSRGSVASLLHGNRGENRIPL 432
Query: 460 DWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPV 519
DW TR+ + +GAA+G+ARIH E + K+ HGN+KSSN+ L+ C+SD GL+ +++P+
Sbjct: 433 DWETRMKIAIGAAKGIARIHKE-NNGKLVHGNIKSSNIFLNSESNGCVSDLGLTAVMSPL 491
Query: 520 -HATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEE 578
+R GYRAPE T+ ++ SQ +DVYSFGV+LLE+LTGK+P
Sbjct: 492 APPISRQAGYRAPEVTDTRKSSQLSDVYSFGVVLLELLTGKSPI-----------HTTAG 540
Query: 579 ETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVD 638
+ ++ L +WV SVVREEWT EVFD ELLRY NIEEE+V ML + ++CVV+ ++RP M D
Sbjct: 541 DEIIHLVRWVHSVVREEWTAEVFDIELLRYTNIEEEMVEMLQIAMSCVVKAADQRPKMSD 600
Query: 639 VVKMIEDI---RVEQSPLCEDYDESRNSLSPSIPTTE 672
+V++IE++ R P E +S N S + +E
Sbjct: 601 LVRLIENVGNRRTSIEPEPELKPKSENGASETSTPSE 637
>At1g48480 protein kinase like protein
Length = 655
Score = 424 bits (1089), Expect = e-118
Identities = 277/664 (41%), Positives = 384/664 (57%), Gaps = 63/664 (9%)
Query: 10 FLFLSIYIVPCLTHNDTQA-LTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRVT 68
FL L + +P + D A T G W + +W GV C +NRVT
Sbjct: 17 FLSLLLLSLPLPSTQDLNADRTALLSLRSAVGGRTFRWNIKQTSPCNWAGVKCE-SNRVT 75
Query: 69 TLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
L LP + L G P +LT LR L L N L+G++ L S +NL+ LYL GN FS
Sbjct: 76 ALRLPGVALSGDIPEGIFGNLTQLRTLSLRLNALSGSLPKDL-STSSNLRHLYLQGNRFS 134
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G+IP + SL++L+RL+L+ N+ G+I + + LT L TL L+NN LSG+IPDL +P
Sbjct: 135 GEIPEVLFSLSHLVRLNLASNSFTGEISSGFTNLTKLKTLFLENNQLSGSIPDLD--LP- 191
Query: 187 LTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVP 246
L + N++NN G +P L +F +SF LCG KP ++C P E V + P
Sbjct: 192 LVQFNVSNNSLNGSIPKN-LQRFESDSFL-QTSLCG-KPLKLC-------PDEETVPSQP 241
Query: 247 SNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCA----RGRG 302
++ + SV + LS G I IV+ V ++ ++ C R R
Sbjct: 242 TSGGNRTPPSVEGSEEKKKKNKLSGGAIAGIVIGCVVGFALIVLILMVLCRKKSNKRSRA 301
Query: 303 VNSNSLMGSEAGKRKSYGSEKKVYNSN------------GGGGDSSDGTSGTDMSKLVFF 350
V+ +++ E + G ++ V N N G G +S+G +G KLVFF
Sbjct: 302 VDISTIKQQEP---EIPGDKEAVDNGNVYSVSAAAAAAMTGNGKASEG-NGPATKKLVFF 357
Query: 351 DRRNG-FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMD 409
F+LEDLLRASAE+LGKG+ GT Y+AVLD + VAVKRLKD A EF++ ++
Sbjct: 358 GNATKVFDLEDLLRASAEVLGKGTFGTAYKAVLDAVTVVAVKRLKDVM-MADKEFKEKIE 416
Query: 410 VIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVL 469
++G + H N+V LRAYY++++EKLLVYD++ GSL ALLHGNRG GR PL+W R + +
Sbjct: 417 LVGAMDHENLVPLRAYYFSRDEKLLVYDFMPMGSLSALLHGNRGAGRSPLNWDVRSRIAI 476
Query: 470 GAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLL-----NPVHATAR 524
GAARGL +H++ HGN+KSSN+LL K+ A +SDFGL+ L+ NP AT
Sbjct: 477 GAARGLDYLHSQ--GTSTSHGNIKSSNILLTKSHDAKVSDFGLAQLVGSSATNPNRAT-- 532
Query: 525 LGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDL 584
GYRAPE T+ KR+SQ+ DVYSFGV+LLE++TGKAPS V EE VDL
Sbjct: 533 --GYRAPEVTDPKRVSQKGDVYSFGVVLLELITGKAPS----------NSVMNEEG-VDL 579
Query: 585 PKWVRSVVREEWTGEVFDQELLRYKNIEEELVS-MLHVGLACVVQQPEKRPTMVDVVKMI 643
P+WV+SV R+EW EVFD ELL EEE+++ M+ +GL C Q P++RP M +VV+ +
Sbjct: 580 PRWVKSVARDEWRREVFDSELLSLATDEEEMMAEMVQLGLECTSQHPDQRPEMSEVVRKM 639
Query: 644 EDIR 647
E++R
Sbjct: 640 ENLR 643
>At3g17840 receptor kinase, putative
Length = 647
Score = 421 bits (1083), Expect = e-118
Identities = 280/665 (42%), Positives = 383/665 (57%), Gaps = 52/665 (7%)
Query: 1 MNYVIMFFFFLFLSIYIVPCLTH--NDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHG 58
M+ + +FF L LS+ + P + D AL FR L W + +W G
Sbjct: 9 MSNLSIFFSILLLSLPL-PSIGDLAADKSALLSFRSAVGGRTLL---WDVKQTSPCNWTG 64
Query: 59 VTCTPNNRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLK 116
V C RVT L LP L G P +LT LR L L N L G++ L +C++L+
Sbjct: 65 VLCD-GGRVTALRLPGETLSGHIPEGIFGNLTQLRTLSLRLNGLTGSLPLDL-GSCSDLR 122
Query: 117 LLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGN 176
LYL GN FSG+IP + SL+NL+RL+L++N +G+I + LT L TL L+NN LSG+
Sbjct: 123 RLYLQGNRFSGEIPEVLFSLSNLVRLNLAENEFSGEISSGFKNLTRLKTLYLENNKLSGS 182
Query: 177 IPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSP 236
+ DL +L + N++NN G +P + L KF +SF G LCG KP VCS E +
Sbjct: 183 LLDLDL---SLDQFNVSNNLLNGSIPKS-LQKFDSDSFVGTS-LCG-KPLVVCS-NEGTV 235
Query: 237 PSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHC 296
PS P + + P T V + K LS G I IV+ V L ++ ++
Sbjct: 236 PSQ------PISVGNIPGT-VEGSEEKKKRKKLSGGAIAGIVIGCVVGLSLIVMILMVLF 288
Query: 297 CARG----RGVNSNSLMGSEAG----KRKSYGSEKKVY-NSNGGGGDSSDGTSGTDMSKL 347
+G R ++ ++ E K E + Y N + + + M KL
Sbjct: 289 RKKGNERTRAIDLATIKHHEVEIPGEKAAVEAPENRSYVNEYSPSAVKAVEVNSSGMKKL 348
Query: 348 VFFDRRNG-FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQ 406
VFF F+LEDLLRASAE+LGKG+ GT Y+AVLD + VAVKRLKD R EF++
Sbjct: 349 VFFGNATKVFDLEDLLRASAEVLGKGTFGTAYKAVLDAVTLVAVKRLKDVTMADR-EFKE 407
Query: 407 YMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRIS 466
++V+G + H N+V LRAYYY+ +EKLLVYD++ GSL ALLHGN+G GR PL+W R
Sbjct: 408 KIEVVGAMDHENLVPLRAYYYSGDEKLLVYDFMPMGSLSALLHGNKGAGRPPLNWEVRSG 467
Query: 467 LVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATA-RL 525
+ LGAARGL +H++ + HGNVKSSN+LL + A +SDFGL+ L++ T R
Sbjct: 468 IALGAARGLDYLHSQDPLSS--HGNVKSSNILLTNSHDARVSDFGLAQLVSASSTTPNRA 525
Query: 526 GGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLP 585
GYRAPE T+ +R+SQ+ADVYSFGV+LLE+LTGKAPS V EE +DL
Sbjct: 526 TGYRAPEVTDPRRVSQKADVYSFGVVLLELLTGKAPS----------NSVMNEEG-MDLA 574
Query: 586 KWVRSVVREEWTGEVFDQELLRYK---NIEEELVSMLHVGLACVVQQPEKRPTMVDVVKM 642
+WV SV REEW EVFD EL+ + ++EEE+ ML +G+ C Q P+KRP MV+VV+
Sbjct: 575 RWVHSVAREEWRNEVFDSELMSIETVVSVEEEMAEMLQLGIDCTEQHPDKRPVMVEVVRR 634
Query: 643 IEDIR 647
I+++R
Sbjct: 635 IQELR 639
>At5g05160 receptor-like protein kinase
Length = 640
Score = 410 bits (1054), Expect = e-114
Identities = 269/664 (40%), Positives = 376/664 (56%), Gaps = 73/664 (10%)
Query: 3 YVIMFFFFLFLSI--YIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVT 60
+V FFFL L+ +V +D QAL F +L NW + +SW G+T
Sbjct: 8 FVAASFFFLLLAATAVLVSADLASDEQALLNFAASVPHPPKL--NWNKNLSLCSSWIGIT 65
Query: 61 C---TPNNRVTTLVLPSLNLRGPID--ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNL 115
C P +RV + LP + L G I L L L++L L +N L GT+ + +LS +L
Sbjct: 66 CDESNPTSRVVAVRLPGVGLYGSIPPATLGKLDALKVLSLRSNSLFGTLPSDILS-LPSL 124
Query: 116 KLLYLAGNDFSGQIP----PEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNN 171
+ LYL N+FSG++ P IS L+ LDLS N+L+G+IP+ + L+ + L LQNN
Sbjct: 125 EYLYLQHNNFSGELTTNSLPSISK--QLVVLDLSYNSLSGNIPSGLRNLSQITVLYLQNN 182
Query: 172 ALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSL 231
+ G I L +P++ +N++ N G +P L K + SF GN LCG P CS
Sbjct: 183 SFDGPIDSLD--LPSVKVVNLSYNNLSGPIPEH-LKKSPEYSFIGNSLLCGP-PLNACSG 238
Query: 232 TENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSF 291
SP S+ P P T + R + K I+AIVV VA+L +
Sbjct: 239 GAISPSSNLPR----------PLTENLHPVRRRQSKAY----IIAIVVGCSVAVLFL-GI 283
Query: 292 VVAHCCAR------GRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMS 345
V C + G G + MG K+ D G + +
Sbjct: 284 VFLVCLVKKTKKEEGGGEGVRTQMGGVNSKKPQ---------------DFGSGVQDPEKN 328
Query: 346 KLVFFDRRN-GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEF 404
KL FF+R N F+LEDLL+ASAE+LGKGS GT Y+AVL+D + V VKRL++ ++ EF
Sbjct: 329 KLFFFERCNHNFDLEDLLKASAEVLGKGSFGTAYKAVLEDTTAVVVKRLREV-VASKKEF 387
Query: 405 EQYMDVIGKL-KHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTT 463
EQ M+++GK+ +H N V L AYYY+K+EKLLVY Y++ GSL ++HGNRG + DW T
Sbjct: 388 EQQMEIVGKINQHSNFVPLLAYYYSKDEKLLVYKYMTKGSLFGIMHGNRGDRGV--DWET 445
Query: 464 RISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATA 523
R+ + G ++ ++ +H+ K HG++KSSN+LL ++ C+SD L L N T
Sbjct: 446 RMKIATGTSKAISYLHS----LKFVHGDIKSSNILLTEDLEPCLSDTSLVTLFNLPTHTP 501
Query: 524 RLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVD 583
R GY APE E +R+SQ++DVYSFGV++LE+LTGK P Q +E+E V+D
Sbjct: 502 RTIGYNAPEVIETRRVSQRSDVYSFGVVILEMLTGKTPLTQ--------PGLEDERVVID 553
Query: 584 LPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMI 643
LP+WVRSVVREEWT EVFD ELL+++NIEEE+V ML + LACV + PE RP M +V +MI
Sbjct: 554 LPRWVRSVVREEWTAEVFDVELLKFQNIEEEMVQMLQLALACVARNPESRPKMEEVARMI 613
Query: 644 EDIR 647
ED+R
Sbjct: 614 EDVR 617
>At5g16590 receptor-like protein kinase
Length = 625
Score = 392 bits (1008), Expect = e-109
Identities = 249/650 (38%), Positives = 369/650 (56%), Gaps = 60/650 (9%)
Query: 9 FFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQ-LLTNWTGPEACSASWHGVTCTPNNRV 67
FF F+ + V D +AL R HG+ LL N T P +W GV C + RV
Sbjct: 12 FFFFICLVSVTSDLEADRRALIALRD--GVHGRPLLWNLTAPPC---TWGGVQCE-SGRV 65
Query: 68 TTLVLPSLNLRGPID-ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
T L LP + L GP+ A+ +LT L L N LNG + +N T L+ LYL GN FS
Sbjct: 66 TALRLPGVGLSGPLPIAIGNLTKLETLSFRFNALNGPLPPDF-ANLTLLRYLYLQGNAFS 124
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G+IP + +L N++R++L+ NN G IP+ ++ T L TL LQ+N L+G IP++
Sbjct: 125 GEIPSFLFTLPNIIRINLAQNNFLGRIPDNVNSATRLATLYLQDNQLTGPIPEIKI---K 181
Query: 187 LTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVP 246
L + N+++N+ G +P+ L+ +F GN LCG KP C P
Sbjct: 182 LQQFNVSSNQLNGSIPDP-LSGMPKTAFLGNL-LCG-KPLDAC----------------P 222
Query: 247 SNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSN 306
N + + + +S LS G IV IV+ V LLV+ F++ C R +
Sbjct: 223 VNGTGNGTVTPGGKGKSDK---LSAGAIVGIVIGCFVLLLVL--FLIVFCLCRKKKKEQV 277
Query: 307 SLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSK--------LVFFDRRNG-FE 357
S S SNG ++G S +SK L FF + G F+
Sbjct: 278 VQSRSIEAAPVPTSSAAVAKESNGPPAVVANGASENGVSKNPAAVSKDLTFFVKSFGEFD 337
Query: 358 LEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHP 417
L+ LL+ASAE+LGKG+ G+ Y+A D G VAVKRL+D EF + + V+G + H
Sbjct: 338 LDGLLKASAEVLGKGTFGSSYKASFDHGLVVAVKRLRDV-VVPEKEFREKLQVLGSISHA 396
Query: 418 NIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLAR 477
N+V L AYY++++EKL+V++Y+S GSL ALLHGN+G GR PL+W TR ++ LGAAR ++
Sbjct: 397 NLVTLIAYYFSRDEKLVVFEYMSRGSLSALLHGNKGSGRSPLNWETRANIALGAARAISY 456
Query: 478 IHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQK 537
+H+ A HGN+KSSN+LL ++ A +SD+ L+ +++P R+ GYRAPE T+ +
Sbjct: 457 LHSR--DATTSHGNIKSSNILLSESFEAKVSDYCLAPMISPTSTPNRIDGYRAPEVTDAR 514
Query: 538 RLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWT 597
++SQ+ADVYSFGVL+LE+LTGK+P+ Q + E VDLP+WV S+ ++
Sbjct: 515 KISQKADVYSFGVLILELLTGKSPTHQ-----------QLHEEGVDLPRWVSSITEQQSP 563
Query: 598 GEVFDQELLRYK-NIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
+VFD EL RY+ + E ++ +L++G++C Q P+ RPTM +V ++IE++
Sbjct: 564 SDVFDPELTRYQSDSNENMIRLLNIGISCTTQYPDSRPTMPEVTRLIEEV 613
>At5g53320 receptor protein kinase-like protein
Length = 601
Score = 392 bits (1006), Expect = e-109
Identities = 254/639 (39%), Positives = 360/639 (55%), Gaps = 99/639 (15%)
Query: 45 NWTGPEACSASWHGVTC-TPNNRVTTLVLPSLNLRGPIDA--LSSLTHLRLLDLHNNRLN 101
NW+ + W GVTC + ++ V L L + LRG I+ ++ L++LR L L +N ++
Sbjct: 44 NWSPSLSICTKWTGVTCNSDHSSVDALHLAATGLRGDIELSIIARLSNLRFLILSSNNIS 103
Query: 102 GTVSASL--LSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISR 159
GT +L L N T LKL + N+FSG +P ++SS L LDLS+N G IP+ I +
Sbjct: 104 GTFPTTLQALKNLTELKLDF---NEFSGPLPSDLSSWERLQVLDLSNNRFNGSIPSSIGK 160
Query: 160 LTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEG 219
LT L +L L N SG IPDL +P L LN+ +N G VP + L +F +F GN+
Sbjct: 161 LTLLHSLNLAYNKFSGEIPDLH--IPGLKLLNLAHNNLTGTVPQS-LQRFPLSAFVGNKV 217
Query: 220 LCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVV 279
L P S + R ++HH + G+ +++
Sbjct: 218 LA-------------------------------PVHSSL-RKHTKHHNHVVLGIALSVCF 245
Query: 280 AICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGT 339
AI +ALL + ++ H R S + +RK SD
Sbjct: 246 AI-LALLAILLVIIIH----NREEQRRSSKDKPSKRRKD-----------------SDPN 283
Query: 340 SGTDMSKLVFFDRRN-GFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANP 398
G +K+VFF+ +N F+LEDLLRASAE+LGKG GT Y+ L+D +T+ VKR+K+ +
Sbjct: 284 VGEGDNKIVFFEGKNLVFDLEDLLRASAEVLGKGPFGTTYKVDLEDSATIVVKRIKEVSV 343
Query: 399 CARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRG-PGRI 457
R EFEQ ++ IG +KH N+ LR Y+Y+K+EKL+VYDY +GSL LLHG +G R
Sbjct: 344 PQR-EFEQQIENIGSIKHENVATLRGYFYSKDEKLVVYDYYEHGSLSTLLHGQKGLRDRK 402
Query: 458 PLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLN 517
L+W TR+++V G ARG+A IH++ S K+ HGN+KSSN+ L+ G CIS G++ L++
Sbjct: 403 RLEWETRLNMVYGTARGVAHIHSQ-SGGKLVHGNIKSSNIFLNGKGYGCISGTGMATLMH 461
Query: 518 --PVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKV 575
P HA GYRAPE T+ ++ +Q +DVYSFG+L+ EVLTGK+
Sbjct: 462 SLPRHAV----GYRAPEITDTRKGTQPSDVYSFGILIFEVLTGKSE-------------- 503
Query: 576 EEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPT 635
V +L +WV SVVREEWTGEVFD+ELLR +EEE+V ML VG+ C + PEKRP
Sbjct: 504 -----VANLVRWVNSVVREEWTGEVFDEELLRCTQVEEEMVEMLQVGMVCTARLPEKRPN 558
Query: 636 MVDVVKMIEDIRVEQSPLCEDYDESRNSLSPSIPTTEDG 674
M++VV+M+E+IR E+ L Y R+ +S TT G
Sbjct: 559 MIEVVRMVEEIRPEK--LASGY---RSEVSTGATTTPIG 592
>At5g24100 receptor-like protein kinase
Length = 614
Score = 387 bits (993), Expect = e-107
Identities = 248/654 (37%), Positives = 361/654 (54%), Gaps = 62/654 (9%)
Query: 3 YVIMFFFFLFLSIYI-VPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTC 61
Y ++F FF ++Y V D QAL F L N + P C+ +W GVTC
Sbjct: 10 YFVLFLFFGSSALYSQVTGDLAGDRQALLDFLNNIIHPRSLAWNTSSP-VCT-TWPGVTC 67
Query: 62 T-PNNRVTTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLL 118
RVT L LP +L G P +S L+ L++L L +N L G L LK +
Sbjct: 68 DIDGTRVTALHLPGASLLGVIPPGTISRLSELQILSLRSNGLRGPFPIDFLQ-LKKLKAI 126
Query: 119 YLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIP 178
L N FSG +P + ++ NL LDL N G IP + LT L++L L N+ SG IP
Sbjct: 127 SLGNNRFSGPLPSDYATWTNLTVLDLYSNRFNGSIPAGFANLTGLVSLNLAKNSFSGEIP 186
Query: 179 DLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPS 238
DL+ +P L LN +NN G +PN+ L +FG+ +FSGN + EN+PP
Sbjct: 187 DLN--LPGLRRLNFSNNNLTGSIPNS-LKRFGNSAFSGNN-----------LVFENAPP- 231
Query: 239 SEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCA 298
PA V + + ++ +S I+ I +++C + V + V+ C
Sbjct: 232 --------------PAV-VSFKEQKKNGIYISEPAILGIAISVCFVIFFVIAVVIIVCYV 276
Query: 299 RGRGVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRN-GFE 357
+ + + + K SEK+V + G + D ++++K++FF+ N F
Sbjct: 277 KRQRKSETEPKPDKLKLAKKMPSEKEV-SKLGKEKNIEDMEDKSEINKVMFFEGSNLAFN 335
Query: 358 LEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHP 417
LEDLL ASAE LGKG G Y+AVL+D +AVKRLKD +R +F+ M+++G +KH
Sbjct: 336 LEDLLIASAEFLGKGVFGMTYKAVLEDSKVIAVKRLKDI-VVSRKDFKHQMEIVGNIKHE 394
Query: 418 NIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHG-NRGPGRIPLDWTTRISLVLGAARGLA 476
N+ LRAY +KEEKL+VYDY SNGSL LHG N G +PL+W TR+ ++G A+GL
Sbjct: 395 NVAPLRAYVCSKEEKLMVYDYDSNGSLSLRLHGKNADEGHVPLNWETRLRFMIGVAKGLG 454
Query: 477 RIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPV---HATAR-LGGYRAPE 532
IHT+ A HGN+KSSNV ++ G CIS+ GL LL NPV ++AR + YRAPE
Sbjct: 455 HIHTQNLA----HGNIKSSNVFMNSEGYGCISEAGLPLLTNPVVRADSSARSVLRYRAPE 510
Query: 533 QTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVV 592
T+ +R + ++D+YSFG+L+LE LTG++ +++ + +DL WV V+
Sbjct: 511 VTDTRRSTPESDIYSFGILMLETLTGRSI-------------MDDRKEGIDLVVWVNDVI 557
Query: 593 REEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
++WTGEVFD EL++ N+E +L+ ML +G +C P KRP MV VV+ +E+I
Sbjct: 558 SKQWTGEVFDLELVKTPNVEAKLLQMLQLGTSCTAMVPAKRPDMVKVVETLEEI 611
>At3g02880 putative protein kinase
Length = 627
Score = 382 bits (982), Expect = e-106
Identities = 241/649 (37%), Positives = 370/649 (56%), Gaps = 58/649 (8%)
Query: 11 LFLSIYIVPCLT---HNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRV 67
+FL ++ + +T +D +AL R LL N + C+ WHGV C RV
Sbjct: 12 VFLFVFYLAAVTSDLESDRRALLAVRNSVRGR-PLLWNMSASSPCN--WHGVHCDAG-RV 67
Query: 68 TTLVLPSLNLRG--PIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDF 125
T L LP L G PI + +LT L+ L L N L+G + + SN L+ LYL GN F
Sbjct: 68 TALRLPGSGLFGSLPIGGIGNLTQLKTLSLRFNSLSGPIPSDF-SNLVLLRYLYLQGNAF 126
Query: 126 SGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMP 185
SG+IP + +L +++R++L +N +G IP+ ++ T L+TL L+ N LSG IP+++ +P
Sbjct: 127 SGEIPSLLFTLPSIIRINLGENKFSGRIPDNVNSATRLVTLYLERNQLSGPIPEIT--LP 184
Query: 186 NLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTV 245
L + N+++N+ G +P++ L+ + +F GN LCG KP C
Sbjct: 185 -LQQFNVSSNQLNGSIPSS-LSSWPRTAFEGNT-LCG-KPLDTCEA-------------- 226
Query: 246 PSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVN- 304
+P+ A P + LS G IV IV+ V LL++ + C R + N
Sbjct: 227 -ESPNGGDAGGPNTPPEKKDSDKLSAGAIVGIVIGCVVGLLLLLLILFCLCRKRKKEENV 285
Query: 305 -SNSLMGSEAGKRKSYGSEKK----VYNSNGGGGDSSDGTSGTDMSKLVFFDRRNG-FEL 358
S ++ A S K+ V + G +S G L FF + G F+L
Sbjct: 286 PSRNVEAPVAAATSSAAIPKETVVVVPPAKATGSES-----GAVNKDLTFFVKSFGEFDL 340
Query: 359 EDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPN 418
+ LL+ASAE+LGKG++G+ Y+A + G VAVKRL+D EF + + V+G + H N
Sbjct: 341 DGLLKASAEVLGKGTVGSSYKASFEHGLVVAVKRLRDV-VVPEKEFRERLHVLGSMSHAN 399
Query: 419 IVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARI 478
+V L AYY++++EKLLV++Y+S GSL A+LHGN+G GR PL+W TR + LGAAR ++ +
Sbjct: 400 LVTLIAYYFSRDEKLLVFEYMSKGSLSAILHGNKGNGRTPLNWETRAGIALGAARAISYL 459
Query: 479 HTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKR 538
H+ HGN+KSSN+LL + A +SD+GL+ +++ A R+ GYRAPE T+ ++
Sbjct: 460 HSRDGTTS--HGNIKSSNILLSDSYEAKVSDYGLAPIISSTSAPNRIDGYRAPEITDARK 517
Query: 539 LSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTG 598
+SQ+ADVYSFGVL+LE+LTGK+P+ Q + E VDLP+WV+SV ++
Sbjct: 518 ISQKADVYSFGVLILELLTGKSPTHQ-----------QLNEEGVDLPRWVQSVTEQQTPS 566
Query: 599 EVFDQELLRYK-NIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
+V D EL RY+ E ++ +L +G++C Q P+ RP+M +V ++IE++
Sbjct: 567 DVLDPELTRYQPEGNENIIRLLKIGMSCTAQFPDSRPSMAEVTRLIEEV 615
>At1g64210 hypothetical protein
Length = 587
Score = 378 bits (970), Expect = e-105
Identities = 240/650 (36%), Positives = 348/650 (52%), Gaps = 76/650 (11%)
Query: 4 VIMFFFFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTP 63
+ +FFF L L ++ T D + L + +L +W SW GVTC
Sbjct: 3 IFLFFFSLILCFVLISSQTLEDDKKALLHFLSSFNSSRL--HWNQSSDVCHSWTGVTCNE 60
Query: 64 N-NRVTTLVLPSLNLRGPIDALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG 122
N +R+ ++ LP++ NG + +S ++LK L L
Sbjct: 61 NGDRIVSVRLPAVGF-----------------------NGLIPPFTISRLSSLKFLSLRK 97
Query: 123 NDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSS 182
N F+G P + ++L +L L L N+L+G + S L NL L L NN +G+IP S
Sbjct: 98 NHFTGDFPSDFTNLKSLTHLYLQHNHLSGPLLAIFSELKNLKVLDLSNNGFNGSIPTSLS 157
Query: 183 IMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPV 242
+ +L LN+ NN F G++PN L K + S N+ L P S +
Sbjct: 158 GLTSLQVLNLANNSFSGEIPNLHLPKLSQINLSNNK------------LIGTIPKSLQRF 205
Query: 243 QTVPSNPSSFPATSVIARPRSQHHK-GLSPGVIVAIVVAICVALLVVTSFVVAHCCARGR 301
Q+ S+F ++ R + + GLS + I+ A CV + SF++ C
Sbjct: 206 QS-----SAFSGNNLTERKKQRKTPFGLSQLAFLLILSAACVLCVSGLSFIMITC----- 255
Query: 302 GVNSNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDMSKLVFFDRRNG-FELED 360
GK + G +K +S+ G +S + + K++FF RN F+L+D
Sbjct: 256 -----------FGKTRISGKLRKRDSSSPPGNWTSRDDNTEEGGKIIFFGGRNHLFDLDD 304
Query: 361 LLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIV 420
LL +SAE+LGKG+ GT Y+ ++D STV VKRLK+ R EFEQ M++IG ++H N+
Sbjct: 305 LLSSSAEVLGKGAFGTTYKVTMEDMSTVVVKRLKEV-VVGRREFEQQMEIIGMIRHENVA 363
Query: 421 KLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGP-GRIPLDWTTRISLVLGAARGLARIH 479
+L+AYYY+K++KL VY Y ++GSL +LHGNRG R+PLDW R+ + GAARGLA+IH
Sbjct: 364 ELKAYYYSKDDKLAVYSYYNHGSLFEILHGNRGRYHRVPLDWDARLRIATGAARGLAKIH 423
Query: 480 TEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARL-GGYRAPEQTEQKR 538
K HGN+KSSN+ LD CI D GL+ ++ + T L GY APE T+ +R
Sbjct: 424 ----EGKFIHGNIKSSNIFLDSQCYGCIGDVGLTTIMRSLPQTTCLTSGYHAPEITDTRR 479
Query: 539 LSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTG 598
+Q +DVYSFGV+LLE+LTGK SP ++ V +DL W+RSVV +EWTG
Sbjct: 480 STQFSDVYSFGVVLLELLTGK-------SPVSQAELVPTGGENMDLASWIRSVVAKEWTG 532
Query: 599 EVFDQELLRYK-NIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
EVFD E+L EEE+V ML +GLACV + ++RP + V+K+IEDIR
Sbjct: 533 EVFDMEILSQSGGFEEEMVEMLQIGLACVALKQQERPHIAQVLKLIEDIR 582
>At1g60630 receptor kinase like protein
Length = 652
Score = 348 bits (892), Expect = 7e-96
Identities = 245/665 (36%), Positives = 362/665 (53%), Gaps = 76/665 (11%)
Query: 9 FFLFLSIYIVPCLTHNDTQALTLFRQQTDTHGQLLTNWTGPEACSASWHGVTCTPNNRVT 68
FFL + +++ + +D +AL + D + W G + C+ W GV RV+
Sbjct: 9 FFLVFAFFLISPVRSSDVEALLSLKSSIDPSNSI--PWRGTDPCN--WEGVKKCMKGRVS 64
Query: 69 TLVLPSLNLRGPID--ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFS 126
LVL +LNL G ++ +L+ L LR+L N L+G++ LS NLK LYL N+FS
Sbjct: 65 KLVLENLNLSGSLNGKSLNQLDQLRVLSFKGNSLSGSIPN--LSGLVNLKSLYLNDNNFS 122
Query: 127 GQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPN 186
G+ P ++SL+ L + LS N +G IP+ + RL+ L T +Q+N SG+IP L+
Sbjct: 123 GEFPESLTSLHRLKTVVLSRNRFSGKIPSSLLRLSRLYTFYVQDNLFSGSIPPLNQA--T 180
Query: 187 LTELNMTNNEFYGKVPNTM-LNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTV 245
L N++NN+ G +P T LN+F + SF+ N LCG + C+ T +
Sbjct: 181 LRFFNVSNNQLSGHIPPTQALNRFNESSFTDNIALCGDQIQNSCNDTTG----------I 230
Query: 246 PSNPSSFPATSVIARPRSQHHK-GLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVN 304
S PS+ PA V A+ RS+ G+ G I ++ + + L++ C R
Sbjct: 231 TSTPSAKPAIPV-AKTRSRTKLIGIISGSICGGILILLLTFLLI--------CLLWRRKR 281
Query: 305 SNSLMGSEAGKRKSYGSEKKVYNSNGGGGD-----------SSDGTSGTDMSKLVFFDRR 353
S S KR + E K + G D S +G+ GT LVF R
Sbjct: 282 SKSKREERRSKRVAESKEAKTAETEEGTSDQKNKRFSWEKESEEGSVGT----LVFLGRD 337
Query: 354 NG---FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDV 410
+ ++DLL+ASAE LG+G+LG+ Y+AV++ G + VKRLKDA EF++++++
Sbjct: 338 ITVVRYTMDDLLKASAETLGRGTLGSTYKAVMESGFIITVKRLKDAGFPRMDEFKRHIEI 397
Query: 411 IGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNR--GPGRIPLDWTTRISLV 468
+G+LKHPN+V LRAY+ AKEE LLVYDY NGSL +L+HG++ G G+ PL WT+ + +
Sbjct: 398 LGRLKHPNLVPLRAYFQAKEECLLVYDYFPNGSLFSLIHGSKVSGSGK-PLHWTSCLKIA 456
Query: 469 LGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPV---HATARL 525
A GL IH + HGN+KSSNVLL + +C++D+GLS L +P +A
Sbjct: 457 EDLAMGLVYIHQN---PGLTHGNLKSSNVLLGPDFESCLTDYGLSDLHDPYSIEDTSAAS 513
Query: 526 GGYRAPEQTEQKRLS-QQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDL 584
Y+APE + ++ S Q ADVYSFGVLLLE+LTG+ K + D+
Sbjct: 514 LFYKAPECRDLRKASTQPADVYSFGVLLLELLTGRTSF-----------KDLVHKYGSDI 562
Query: 585 PKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIE 644
WVR+ VREE T EV ++ EE+L ++L + ACV +PE RP M +V+KM++
Sbjct: 563 STWVRA-VREEET-EVSEE----LNASEEKLQALLTIATACVAVKPENRPAMREVLKMVK 616
Query: 645 DIRVE 649
D R E
Sbjct: 617 DARAE 621
>At3g51740 unknown protein
Length = 836
Score = 330 bits (847), Expect = 1e-90
Identities = 218/570 (38%), Positives = 309/570 (53%), Gaps = 51/570 (8%)
Query: 87 LTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSD 146
L HL+ LD N +NGT+ S SN ++L L L N G IP I L+NL L+L
Sbjct: 286 LPHLQSLDFSYNSINGTIPDSF-SNLSSLVSLNLESNHLKGPIPDAIDRLHNLTELNLKR 344
Query: 147 NNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTML 206
N + G IP I ++ + L L N +G IP + L+ N++ N G VP +
Sbjct: 345 NKINGPIPETIGNISGIKKLDLSENNFTGPIPLSLVHLAKLSSFNVSYNTLSGPVPPVLS 404
Query: 207 NKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHH 266
KF SF GN LCG C +P P+ P++ PR HH
Sbjct: 405 KKFNSSSFLGNIQLCGYSSSNPCP----APDHHHPLTLSPTSSQE---------PRKHHH 451
Query: 267 KGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVNSNSLMGSEAGKRKSYGSEKKVY 326
+ LS ++ I + +A+L++ ++ C + R + + GK K+ SEK V
Sbjct: 452 RKLSVKDVILIAIGALLAILLLLCCILLCCLIKKRAA-----LKQKDGKDKT--SEKTV- 503
Query: 327 NSNGGGGDSSDGTSGTDMSKLVFFDRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGS 386
S G G +S G G KLV FD F +DLL A+AE++GK + GT Y+A L+DG+
Sbjct: 504 -SAGVAGTASAG--GEMGGKLVHFDGPFVFTADDLLCATAEIMGKSTYGTAYKATLEDGN 560
Query: 387 TVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYA-KEEKLLVYDYLSNGSLH 445
VAVKRL++ EFE + +GK++H N++ LRAYY K EKLLV+DY+S GSL
Sbjct: 561 EVAVKRLREKTTKGVKEFEGEVTALGKIRHQNLLALRAYYLGPKGEKLLVFDYMSKGSLS 620
Query: 446 ALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVA 505
A LH RGP + + W TR+ + G +RGLA +H S + H N+ +SN+LLD+ A
Sbjct: 621 AFLHA-RGPETL-IPWETRMKIAKGISRGLAHLH---SNENMIHENLTASNILLDEQTNA 675
Query: 506 CISDFGLSLLLNP-----VHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKA 560
I+D+GLS L+ V ATA GYRAPE ++ K S + DVYS G+++LE+LTGK+
Sbjct: 676 HIADYGLSRLMTAAAATNVIATAGTLGYRAPEFSKIKNASAKTDVYSLGIIILELLTGKS 735
Query: 561 PSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLR-YKNIEEELVSML 619
P E +DLP+WV S+V+EEWT EVFD EL+R +++ +EL++ L
Sbjct: 736 PG--------------EPTNGMDLPQWVASIVKEEWTNEVFDLELMRETQSVGDELLNTL 781
Query: 620 HVGLACVVQQPEKRPTMVDVVKMIEDIRVE 649
+ L CV P RP VV+ +E+IR E
Sbjct: 782 KLALHCVDPSPAARPEANQVVEQLEEIRPE 811
Score = 92.0 bits (227), Expect = 9e-19
Identities = 89/287 (31%), Positives = 127/287 (44%), Gaps = 37/287 (12%)
Query: 21 LTHNDTQALTLFRQQTDTHGQLLTNWTGP---EACSASWHGVTCTPNNRVTTLVLPSLNL 77
+T + QAL + + +L +W + CS W G+ C +V + LP L
Sbjct: 49 VTQANYQALQAIKHELIDFTGVLKSWNNSASSQVCSG-WAGIKCL-RGQVVAIQLPWKGL 106
Query: 78 RGPI-DALSSLTHLRLLDLHNNRLNGTVSASL-----------------------LSNCT 113
G I + + L LR L LHNN + G+V SL L NC
Sbjct: 107 GGTISEKIGQLGSLRKLSLHNNVIAGSVPRSLGYLKSLRGVYLFNNRLSGSIPVSLGNCP 166
Query: 114 NLKLLYLAGNDFSGQIPPEISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNAL 173
L+ L L+ N +G IPP ++ L RL+LS N+L+G +P ++R L L LQ+N L
Sbjct: 167 LLQNLDLSSNQLTGAIPPSLTESTRLYRLNLSFNSLSGPLPVSVARSYTLTFLDLQHNNL 226
Query: 174 SGNIPD--LSSIMPNLTELNMTNNEFYGKVPNTML--NKFGDESFSGNEGLCGSKPFQVC 229
SG+IPD ++ P L LN+ +N F G VP ++ + + S S N+ L GS P +
Sbjct: 227 SGSIPDFFVNGSHP-LKTLNLDHNRFSGAVPVSLCKHSLLEEVSISHNQ-LSGSIPRECG 284
Query: 230 SLTENSPPSSEPVQTVPSNPSSFPATS--VIARPRSQHHKGLSPGVI 274
L + P SF S V S H KG P I
Sbjct: 285 GLPHLQSLDFSYNSINGTIPDSFSNLSSLVSLNLESNHLKGPIPDAI 331
>At1g50610 receptor-like protein kinase, putative
Length = 686
Score = 323 bits (827), Expect = 3e-88
Identities = 220/665 (33%), Positives = 347/665 (52%), Gaps = 57/665 (8%)
Query: 8 FFFLFLSIYIVPCLTH-----NDTQALTLFRQQTDTHGQLLTNW---TGP-EACSASWHG 58
F +F+S+ ++ +D L F+ T +G +W + P + +A+W G
Sbjct: 25 FIIIFISVLCPVAMSQVVVPDSDADCLLRFKD-TLANGSEFRSWDPLSSPCQGNTANWFG 83
Query: 59 VTCTPNNRVTTLVLPSLNLRGPI--DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLK 116
V C+ N V L L + L G + D L + +LR + NN NG + + T+LK
Sbjct: 84 VLCS--NYVWGLQLEGMGLTGKLNLDPLVPMKNLRTISFMNNNFNGPMPQ--VKRFTSLK 139
Query: 117 LLYLAGNDFSGQIPPE-ISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSG 175
LYL+ N FSG+IP + + L ++ L++N G IP+ ++ L LL LRL N G
Sbjct: 140 SLYLSNNRFSGEIPADAFLGMPLLKKILLANNAFRGTIPSSLASLPMLLELRLNGNQFQG 199
Query: 176 NIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENS 235
IP +L + NN+ G +P ++ N SF+GN+GLC + P CS
Sbjct: 200 QIPSFQQ--KDLKLASFENNDLDGPIPESLRN-MDPGSFAGNKGLCDA-PLSPCS----- 250
Query: 236 PPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAH 295
SS V VP +P +TS G G + + + V +++ +
Sbjct: 251 -SSSPGVPVVPVSPVDPKSTSPPT--------GKKAGSFYTLAIILIVIGIILVIIALVF 301
Query: 296 CCARGRGVNSNSLMGSEAGKRK--SYGSEKKVYNSNGGGGDSSDGTSGTDM----SKLVF 349
C + R N S S AGK + SY + N N +S + T M +L+F
Sbjct: 302 CFVQSRRRNFLSAYPSSAGKERIESYNYHQST-NKNNKPAESVNHTRRGSMPDPGGRLLF 360
Query: 350 F-DRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYM 408
D F+L+DLLRASAE+LG G+ G Y+A + G T+ VKR K N R EF ++M
Sbjct: 361 VRDDIQRFDLQDLLRASAEVLGSGTFGASYKAAISSGQTLVVKRYKHMNNVGRDEFHEHM 420
Query: 409 DVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLV 468
+G+L HPNI+ L AYYY +EEKLLV +++ N SL + LH N G LDW TR+ ++
Sbjct: 421 RRLGRLNHPNILPLVAYYYRREEKLLVTEFMPNSSLASHLHANNSAG---LDWITRLKII 477
Query: 469 LGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGY 528
G A+GL+ + E +PHG++KSSN++LD + ++D+ L +++ HA + Y
Sbjct: 478 KGVAKGLSYLFDELPTLTIPHGHMKSSNIVLDDSFEPLLTDYALRPMMSSEHAHNFMTAY 537
Query: 529 RAPEQTEQKR--LSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPK 586
++PE K ++++ DV+ FGVL+LEVLTG+ P + ++ + L
Sbjct: 538 KSPEYRPSKGQIITKKTDVWCFGVLILEVLTGRFPENYL---------TQGYDSNMSLVT 588
Query: 587 WVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDI 646
WV +V+E+ TG+VFD+E+ KN + E++++L +GL C ++ E+R M +VV+M+E +
Sbjct: 589 WVNDMVKEKKTGDVFDKEMKGKKNCKAEMINLLKIGLRCCEEEEERRMDMREVVEMVEML 648
Query: 647 RVEQS 651
R +S
Sbjct: 649 REGES 653
>At2g07040 putative leucine-rich repeat transmembrane protein kinase
Length = 647
Score = 322 bits (826), Expect = 3e-88
Identities = 222/657 (33%), Positives = 329/657 (49%), Gaps = 49/657 (7%)
Query: 9 FFLFLSIYIVPCLTHNDTQALTLFRQQTDT-HGQLLTNWTGPEACSASWHGVTCTPNNRV 67
F +S++ + ++T+ L F+ L +W W GV C V
Sbjct: 8 FVSIVSVFFMVVNGVSETETLLKFKNSLVIGRANALESWNRRNP-PCKWTGVLCD-RGFV 65
Query: 68 TTLVLPSLNLRGPID--ALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAGNDF 125
L L +L L G ID AL L LR L NN+ G LK LYL+ N F
Sbjct: 66 WGLRLENLELSGSIDIEALMGLNSLRSLSFINNKFKGPFPE--FKKLVALKSLYLSNNQF 123
Query: 126 SGQIPPE-ISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLSSIM 184
+IP + + L +L L NN G+IP + + L+ LRL N +G IP+
Sbjct: 124 DLEIPKDAFDGMGWLKKLHLEQNNFIGEIPTSLVKSPKLIELRLDGNRFTGQIPEFRH-H 182
Query: 185 PNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVCSLTENSPPSSEPVQT 244
PN+ LN++NN G++PN+ + + F GN+GLCG CS N SSEP +
Sbjct: 183 PNM--LNLSNNALAGQIPNSF-STMDPKLFEGNKGLCGKPLDTKCSSPYNH--SSEPKSS 237
Query: 245 VPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCARGRGVN 304
S F IVA VA A L++ V+ R +
Sbjct: 238 TKKTSSKFL-------------------YIVAAAVAALAASLIIIGVVIFLIRRRKK--- 275
Query: 305 SNSLMGSEAGKRKSYGSEKKVYNSNGGGGDSSDGTSGTDM---SKLVFF-DRRNGFELED 360
L+ +E G + G G S + M +KL F D + FEL+D
Sbjct: 276 KQPLLSAEPGPSSLQMRAGIQESERGQGSYHSQNRAAKKMIHTTKLSFLRDDKGKFELQD 335
Query: 361 LLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQYMDVIGKLKHPNIV 420
LL+ASAE+LG G G Y+ +L +GS + VKR K N EF+++M +G+L H N++
Sbjct: 336 LLKASAEILGSGCFGASYKTLLSNGSVMVVKRFKHMNSAGIDEFQEHMKRLGRLNHENLL 395
Query: 421 KLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISLVLGAARGLARIHT 480
+ AYYY KEEKL V D+++NGSL A LHG++ G+ LDW TR ++V G RGL +H
Sbjct: 396 PIVAYYYKKEEKLFVSDFVANGSLAAHLHGHKSLGQPSLDWPTRFNIVKGVGRGLLYLHK 455
Query: 481 EYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGGYRAPEQTEQKRLS 540
+ PHG++KSSNVLL + + D+GL ++N A + Y++PE +Q R++
Sbjct: 456 NLPSLMAPHGHLKSSNVLLSEKFEPLLMDYGLIPMINEESAQELMVAYKSPEYVKQSRVT 515
Query: 541 QQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKWVRSVVREEWTGEV 600
++ DV+ GVL+LE+LTGK L+ S ++ E EE DL WVRS + EWT E+
Sbjct: 516 KKTDVWGLGVLILEILTGKL--LESFSQVDK----ESEE---DLASWVRSSFKGEWTQEL 566
Query: 601 FDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIRVEQSPLCEDY 657
FDQE+ + N E +++++ +GL+C EKR + + V+ +ED+ E+ +D+
Sbjct: 567 FDQEMGKTSNCEAHILNLMRIGLSCCEVDVEKRLDIREAVEKMEDLMKEREQGDDDF 623
>At5g20690 receptor protein kinase - like
Length = 659
Score = 319 bits (818), Expect = 3e-87
Identities = 224/679 (32%), Positives = 340/679 (49%), Gaps = 69/679 (10%)
Query: 7 FFFFLFLSIYIVPCLTH-NDTQALTLFRQQTDTHGQLLTNWT-GPEACSASWHGVTCTPN 64
F L LS I P L + ++++ L F+ L +W G + CS W G+ C
Sbjct: 11 FLLILLLSFSISPSLQYVSESEPLVRFKNSVKITKGDLNSWREGTDPCSGKWFGIYCQKG 70
Query: 65 NRVTTLVLPSLNLRGPI--DALSSLTHLRLLDLHNNRLNGTVSASLLSNCTNLKLLYLAG 122
V+ + + L L G I D L L +L+ + L NN L+G + LK L L+
Sbjct: 71 LTVSGIHVTRLGLSGTITVDDLKDLPNLKTIRLDNNLLSGPLPHFF--KLRGLKSLMLSN 128
Query: 123 NDFSGQIPPEI-SSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQNNALSGNIPDLS 181
N FSG+I + ++ L RL L N G IP+ I++L L L +Q+N L+G IP
Sbjct: 129 NSFSGEIRDDFFKDMSKLKRLFLDHNKFEGSIPSSITQLPQLEELHMQSNNLTGEIPPEF 188
Query: 182 SIMPNLTELNMTNNEFYGKVPNTMLNKFG-DESFSGNEGLCGSKPFQVCSLTE-NSPPSS 239
M NL L+++ N G VP ++ +K + + NE LCG C E N P
Sbjct: 189 GSMKNLKVLDLSTNSLDGIVPQSIADKKNLAVNLTENEYLCGPVVDVGCENIELNDPQEG 248
Query: 240 EPVQTVPSNPSS-FPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVTSFVVAHCCA 298
+P PS PSS P TS + I AI+V+I +LL++ +V
Sbjct: 249 QP----PSKPSSSVPETS-------------NKAAINAIMVSI--SLLLLFFIIVGVIKR 289
Query: 299 RGRGVNSNSLM-----------------GSEAGKRKSYGSEKK------------VYNSN 329
R + N + M S KR + S K+ V N
Sbjct: 290 RNKKKNPDFRMLANNRENDVVEVRISESSSTTAKRSTDSSRKRGGHSDDGSTKKGVSNIG 349
Query: 330 GGGGDSSDGTSGTDMSKLVFFDRRNG-FELEDLLRASAEMLGKGSLGTVYRAVLDDGSTV 388
GG G G M ++ + G F L DL++A+AE+LG GSL + Y+AV+ G +V
Sbjct: 350 KGGNGGGGGALGGGMGDIIMVNTDKGSFGLPDLMKAAAEVLGNGSLRSAYKAVMTTGLSV 409
Query: 389 AVKRLKDANPCARHEFEQYMDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALL 448
VKR++D N AR F+ M GKL+HPNI+ AY+Y +EEKL+V +Y+ SL +L
Sbjct: 410 VVKRIRDMNQLAREPFDVEMRRFGKLRHPNILTPLAYHYRREEKLVVSEYMPKSSLLYVL 469
Query: 449 HGNRGPGRIPLDWTTRISLVLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACIS 508
HG+RG L W TR+ ++ G A G+ +H E+++ +PHGN+KSSNVLL + IS
Sbjct: 470 HGDRGIYHSELTWATRLKIIQGVAHGMKFLHEEFASYDLPHGNLKSSNVLLSETYEPLIS 529
Query: 509 DFGLSLLLNPVHATARLGGYRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSP 568
D+ LL P +A+ L ++ PE + +++S ++DVY G+++LE+LTGK PS QY
Sbjct: 530 DYAFLPLLQPSNASQALFAFKTPEFAQTQQVSHKSDVYCLGIIILEILTGKFPS-QY--- 585
Query: 569 ANRPRKVEEEETVVDLPKWVRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQ 628
+ + D+ +WV+S V E+ E+ D E++ ++V +L VG AC+
Sbjct: 586 ------LNNGKGGTDIVQWVQSSVAEQKEEELIDPEIVNNTESMRQMVELLRVGAACIAS 639
Query: 629 QPEKRPTMVDVVKMIEDIR 647
P++R M + V+ IE ++
Sbjct: 640 NPDERLDMREAVRRIEQVK 658
>At3g20190 receptor kinase like protein
Length = 679
Score = 319 bits (817), Expect = 4e-87
Identities = 202/620 (32%), Positives = 342/620 (54%), Gaps = 42/620 (6%)
Query: 53 SASWHGVTCTPNNRVTTLVLPSLNLRGPIDA--LSSLTHLRLLDLHNNRLNGTVSASLLS 110
S +W GV C N V L L + L G +D L+++ +LR L NN+ NG++ + +
Sbjct: 83 SENWFGVLCVTGN-VWGLQLEGMGLTGKLDLEPLAAIKNLRTLSFMNNKFNGSMPS--VK 139
Query: 111 NCTNLKLLYLAGNDFSGQIPPE-ISSLNNLLRLDLSDNNLAGDIPNEISRLTNLLTLRLQ 169
N LK LYL+ N F+G+IP + +++L +L L++N G IP+ ++ L LL LRL
Sbjct: 140 NFGALKSLYLSNNRFTGEIPADAFDGMHHLKKLLLANNAFRGSIPSSLAYLPMLLELRLN 199
Query: 170 NNALSGNIPDLSSIMPNLTELNMTNNEFYGKVPNTMLNKFGDESFSGNEGLCGSKPFQVC 229
N G IP +L + NN+ G +P ++ N SFSGN+ LCG P C
Sbjct: 200 GNQFHGEIPYFKQ--KDLKLASFENNDLEGPIPESLSN-MDPVSFSGNKNLCGP-PLSPC 255
Query: 230 SLTENSPPSSEPVQTVPSNPSSFPATSVIARPRSQHHKGLSPGVIVAIVVAICVALLVVT 289
S S P +PS+P+ + +K S +I +++ I + L++++
Sbjct: 256 SSDSGSSPD------LPSSPT-------------EKNKNQSFFIIAIVLIVIGIILMIIS 296
Query: 290 SFVVAHCCARGRGVNSNSLMGSEAGKRKSYG-SEKKVYNSNGGGGDSSDGTSGTDMSKLV 348
V R + +++ G + ++ +Y S K ++ +S + D +KL+
Sbjct: 297 LVVCILHTRRRKSLSAYPSAGQDRTEKYNYDQSTDKDKAADSVTSYTSRRGAVPDQNKLL 356
Query: 349 FF-DRRNGFELEDLLRASAEMLGKGSLGTVYRAVLDDGSTVAVKRLKDANPCARHEFEQY 407
F D F+L+DLLRASAE+LG GS G+ Y+ ++ G + VKR K N R EF ++
Sbjct: 357 FLQDDIQRFDLQDLLRASAEVLGSGSFGSSYKTGINSGQMLVVKRYKHMNNVGRDEFHEH 416
Query: 408 MDVIGKLKHPNIVKLRAYYYAKEEKLLVYDYLSNGSLHALLHGNRGPGRIPLDWTTRISL 467
M +G+LKHPN++ + AYYY +EEKLL+ +++ N SL + LH N + LDW TR+ +
Sbjct: 417 MRRLGRLKHPNLLPIVAYYYRREEKLLIAEFMPNRSLASHLHANHSVDQPGLDWPTRLKI 476
Query: 468 VLGAARGLARIHTEYSAAKVPHGNVKSSNVLLDKNGVACISDFGLSLLLNPVHATARLGG 527
+ G A+GL + E + +PHG++KSSNV+LD++ ++D+ L ++N + +
Sbjct: 477 IQGVAKGLGYLFNELTTLTIPHGHLKSSNVVLDESFEPLLTDYALRPVMNSEQSHNLMIS 536
Query: 528 YRAPEQTEQKRLSQQADVYSFGVLLLEVLTGKAPSLQYPSPANRPRKVEEEETVVDLPKW 587
Y++PE + + L+++ DV+ GVL+LE+LTG+ P Y S + + + L W
Sbjct: 537 YKSPEYSLKGHLTKKTDVWCLGVLILELLTGRFPE-NYLS--------QGYDANMSLVTW 587
Query: 588 VRSVVREEWTGEVFDQELLRYKNIEEELVSMLHVGLACVVQQPEKRPTMVDVVKMIEDIR 647
V ++V+E+ TG+VFD+E+ KN + E++++L +GL+C + E+R M D V+ IE R
Sbjct: 588 VSNMVKEKKTGDVFDKEMTGKKNCKAEMLNLLKIGLSCCEEDEERRMEMRDAVEKIE--R 645
Query: 648 VEQSPLCEDYDESRNSLSPS 667
+++ D+ + +++ S
Sbjct: 646 LKEGEFDNDFASTTHNVFAS 665
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,039,960
Number of Sequences: 26719
Number of extensions: 741874
Number of successful extensions: 12662
Number of sequences better than 10.0: 1208
Number of HSP's better than 10.0 without gapping: 932
Number of HSP's successfully gapped in prelim test: 277
Number of HSP's that attempted gapping in prelim test: 2623
Number of HSP's gapped (non-prelim): 3395
length of query: 676
length of database: 11,318,596
effective HSP length: 106
effective length of query: 570
effective length of database: 8,486,382
effective search space: 4837237740
effective search space used: 4837237740
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC139525.8


