

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.4 + phase: 0
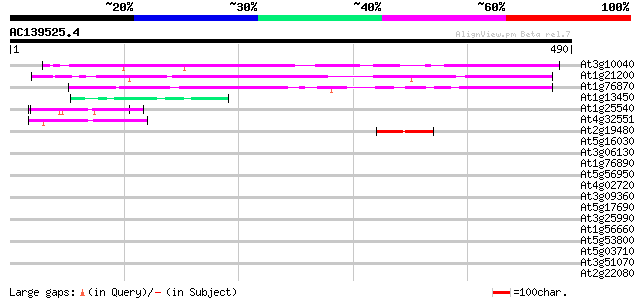
(490 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g10040 unknown protein 382 e-106
At1g21200 unknown protein 272 3e-73
At1g76870 hypothetical protein 237 1e-62
At1g13450 hypothetical protein 47 3e-05
At1g25540 hypothetical protein 45 7e-05
At4g32551 Leunig protein 43 5e-04
At2g19480 putative nucleosome assembly protein 43 5e-04
At5g16030 unknown protein 42 0.001
At3g06130 unknown protein 42 0.001
At1g76890 unknown protein 41 0.002
At5g56950 nucleosome assembly protein 40 0.002
At4g02720 unknown protein 40 0.002
At3g09360 putative transcription factor 40 0.002
At5g17690 chromo domain protein 40 0.003
At3g25990 putative DNA-binding protein, GT-1 40 0.003
At1g56660 hypothetical protein 40 0.003
At5g53800 unknown protein 40 0.004
At5g03710 putative protein 40 0.004
At3g51070 putative protein 40 0.004
At2g22080 En/Spm-like transposon protein 40 0.004
>At3g10040 unknown protein
Length = 418
Score = 382 bits (981), Expect = e-106
Identities = 224/467 (47%), Positives = 276/467 (58%), Gaps = 84/467 (17%)
Query: 29 QNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNKQQQQSSQM 88
QN PN Q N Q H P + T +Q +K +P S K
Sbjct: 8 QNPPNPQ--NSIQFQH-------PHPYTTSGDQQTQPPIKSLYPYASKPKQMSPISGGGC 58
Query: 89 SDEDEPNFPA-----EESSGGDPKRKISPWQRMKWTDTMVRLLIMAVYYIGDEAGSEGTD 143
DED + E+S+G D KRK+S W RMKWTDTMVRLLIMAV+YIGDEAG
Sbjct: 59 DDEDRGSGSGSGCNPEDSAGTDGKRKLSQWHRMKWTDTMVRLLIMAVFYIGDEAGLNDPV 118
Query: 144 PNKKKSSG---------LLQKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVND 194
KKK+ G +LQKKGKWKSVSRAM+EKGF VSPQQCEDKFNDLNKRYKRVND
Sbjct: 119 DAKKKTGGGGGGGGGGGMLQKKGKWKSVSRAMVEKGFSVSPQQCEDKFNDLNKRYKRVND 178
Query: 195 ILGKGTACRVVENQGLLDSMD-LSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATS 253
ILGKG ACRVVENQGLL+SMD L+PK+KDEV+KLLNSKHLFFREMCAYHNSCGH G
Sbjct: 179 ILGKGIACRVVENQGLLESMDHLTPKLKDEVKKLLNSKHLFFREMCAYHNSCGHLG---- 234
Query: 254 NVQHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKI 313
Q PQQ QQ+CFH++E G + R E
Sbjct: 235 -------------GHDQQPPQQNPISIPIPSQQQNCFHAAEAGKMARIAERVE------- 274
Query: 314 GSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFS 373
VEEE + D ++SE +E E+E +RK+ R
Sbjct: 275 ----VEEEVESDMAEDSESEMEESEEEE---------------------TRKKRR----- 304
Query: 374 FPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQ 433
+ V ++ E + V +D GKS WEKK+WIR +++++EE+KIGYE + ++EKQ
Sbjct: 305 ------ISTAVKRLREEAASVVEDVGKSVWEKKEWIRRKMLEIEEKKIGYEWEGVEMEKQ 358
Query: 434 RLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKELELMHIQ 480
R+KW RY SKKEREME+AKL+N+RRRLE ERM+L++R+ E+EL +Q
Sbjct: 359 RVKWMRYRSKKEREMEKAKLDNQRRRLETERMILMLRRSEIELNELQ 405
>At1g21200 unknown protein
Length = 443
Score = 272 bits (696), Expect = 3e-73
Identities = 168/472 (35%), Positives = 252/472 (52%), Gaps = 80/472 (16%)
Query: 20 HQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKN 79
HQ Q+ ++ PN + + L + V HH Q+ + SM S +
Sbjct: 31 HQDSMNQQHRHNPNSRPLH-EGLPFTMVTGQTCDHH-----QNQNMSM--------SEQQ 76
Query: 80 KQQQQSSQMSDEDEPNFPAEESSG----GDPKRKISPWQRMKWTDTMVRLLIMAVYYIGD 135
K +++ + +SD+DEP+F E G + K SPWQR+KWTD MV+LLI AV YIGD
Sbjct: 77 KAEREKNSVSDDDEPSFTEEGGDGVHNEANRSTKGSPWQRVKWTDKMVKLLITAVSYIGD 136
Query: 136 EAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVNDI 195
++ D + ++ +LQKKGKWKSVS+ M E+G++VSPQQCEDKFNDLNKRYK++ND+
Sbjct: 137 DSS---IDSSSRRKFAVLQKKGKWKSVSKVMAERGYHVSPQQCEDKFNDLNKRYKKLNDM 193
Query: 196 LGKGTACRVVENQGLLDSMD-LSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSN 254
LG+GT+C+VVEN LLDS+ L+ K KD+VRK+++SKHLF+ EMC+YHN
Sbjct: 194 LGRGTSCQVVENPALLDSIGYLNDKEKDDVRKIMSSKHLFYEEMCSYHNGNRLHLPHDLA 253
Query: 255 VQHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIG 314
+Q ++ + N ++HQ
Sbjct: 254 LQRSLQLALRSRDDHDNDDSRKHQ------------------------------------ 277
Query: 315 SGYVEEEDDEDEEDESEDFSDEGEDESGEG-CSKGH----------INDQDEEENDGKPS 363
+E+ DDED + + ++ + E G C H I E+ PS
Sbjct: 278 ---MEDLDDEDHDGDGDEHDEYEEQHYAYGDCRVNHYGGGGGPLKKIRPSLSHEDGDHPS 334
Query: 364 RKRARK-GGFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIG 422
+ + S P+ S VNQ E G++ +KQW+ +R +QLEEQK+
Sbjct: 335 HVNSLECNKVSLPQIPFSQADVNQGGAE-------SGRAGSVQKQWMESRTLQLEEQKLQ 387
Query: 423 YESQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKEL 474
+ + +LEKQR +W R+S K+++E+ER ++ENER +LEN+RM L ++++EL
Sbjct: 388 IQVELLELEKQRFRWQRFSKKRDQELERMRMENERMKLENDRMGLELKQREL 439
>At1g76870 hypothetical protein
Length = 385
Score = 237 bits (605), Expect = 1e-62
Identities = 148/432 (34%), Positives = 232/432 (53%), Gaps = 81/432 (18%)
Query: 52 PQHHDTDTNQHHHHSMKH-GFPPFSSSK-NKQQQQSSQMSDEDEPNFPAEESSGGDPKRK 109
P + + QHH +S + GF ++ N + MS++DE S G + ++
Sbjct: 22 PNAINQNQKQHHPNSRQDSGFNNTMDTRHNNVDRGKKSMSEDDE--LCLLSSDGQNKSKE 79
Query: 110 ISPWQRMKWTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEK 169
SPWQR+KW D MV+L+I A+ YIG+++GS+ K +LQKKGKW+SVS+ M E+
Sbjct: 80 NSPWQRVKWMDKMVKLMITALSYIGEDSGSD-------KKFAVLQKKGKWRSVSKVMDER 132
Query: 170 GFYVSPQQCEDKFNDLNKRYKRVNDILGKGTACRVVENQGLLDSMD-LSPKMKDEVRKLL 228
G++VSPQQCEDKFNDLNKRYK++N++LG+GT+C VVEN LLD +D L+ K KDEVR+++
Sbjct: 133 GYHVSPQQCEDKFNDLNKRYKKLNEMLGRGTSCEVVENPSLLDKIDYLNEKEKDEVRRIM 192
Query: 229 NSKHLFFREMCAYHNSCGHGGVATSNVQHQVEGGSGTTTPPQNQPQQQHQH------QHQ 282
+SKHLF+ EMC+YHN N H P + Q+ H +
Sbjct: 193 SSKHLFYEEMCSYHN---------GNRLHL----------PHDPAVQRSLHLITLGSRDD 233
Query: 283 QQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDESEDFSDEGEDESG 342
N +H H +E+ ++DD+ EED SD
Sbjct: 234 HDNDEHGKHQNED-----------------------LDDDDDYEEDHDGALSDR------ 264
Query: 343 EGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSSSSTQLVNQMNNEISGVFQDGGKST 402
+ E+ G P++ G+ P S VN+ G+ D K+
Sbjct: 265 ---PLKRLRQSQSHEDVGHPNK------GYDVPCLPRSQADVNR------GISLDSRKAA 309
Query: 403 WEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLEN 462
++Q I ++ ++LE +K+ +++ +LE+Q+ KW +S ++E+++ + ++ENER +LEN
Sbjct: 310 GLQRQQIESKSLELEGRKLQIQAEMMELERQQFKWEVFSKRREQKLAKMRMENERMKLEN 369
Query: 463 ERMVLLIRKKEL 474
ERM L +++ EL
Sbjct: 370 ERMSLELKRIEL 381
>At1g13450 hypothetical protein
Length = 406
Score = 46.6 bits (109), Expect = 3e-05
Identities = 38/138 (27%), Positives = 52/138 (37%), Gaps = 27/138 (19%)
Query: 54 HHDTDTNQHHHHSMKHGFPPFSSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKRKISPW 113
+ D HHHH+ N QS + + ESSG D + K
Sbjct: 38 NESVDLQSHHHHN----------HHNHHLHQS-----QPQQQILLGESSGEDHEVKAPKK 82
Query: 114 QRMKWTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEKGFYV 173
+ W R LIM G +G K + L W+ +S M EKGF
Sbjct: 83 RAETWVQDETRSLIMF------RRGMDGLFNTSKSNKHL------WEQISSKMREKGFDR 130
Query: 174 SPQQCEDKFNDLNKRYKR 191
SP C DK+ +L K +K+
Sbjct: 131 SPTMCTDKWRNLLKEFKK 148
>At1g25540 hypothetical protein
Length = 809
Score = 45.4 bits (106), Expect = 7e-05
Identities = 34/100 (34%), Positives = 43/100 (43%), Gaps = 17/100 (17%)
Query: 17 IPLHQQQQQQKQQNLPNQQQQNPHQL----HH---SQVVSYAPQHHDTDTNQHHHHSMKH 69
+P QQQQQQ Q QQQQ HQL HH Q Q H QHHH +
Sbjct: 687 MPQLQQQQQQHQ-----QQQQQQHQLSQLQHHQQQQQQQQQQQQQHQLTQLQHHHQQQQQ 741
Query: 70 GFP-----PFSSSKNKQQQQSSQMSDEDEPNFPAEESSGG 104
P +S N+ QQQ+S ++ + P + GG
Sbjct: 742 ASPLNQMQQQTSPLNQMQQQTSPLNQMQQQQQPQQMVMGG 781
Score = 44.7 bits (104), Expect = 1e-04
Identities = 29/100 (29%), Positives = 47/100 (47%), Gaps = 3/100 (3%)
Query: 19 LHQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSK 78
LHQQQQQQ+Q QQQQ+ Q Q+ QH QH ++H +
Sbjct: 662 LHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQH--HQQQQQQ 719
Query: 79 NKQQQQSSQMSDEDEPNFPAEESSG-GDPKRKISPWQRMK 117
+QQQQ Q++ + +++S +++ SP +M+
Sbjct: 720 QQQQQQQHQLTQLQHHHQQQQQASPLNQMQQQTSPLNQMQ 759
Score = 38.1 bits (87), Expect = 0.011
Identities = 25/93 (26%), Positives = 37/93 (38%), Gaps = 1/93 (1%)
Query: 1 MFSTITPSGLLGVVDNIPLHQQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTN 60
+ + P ++ IP +QQQQQQ+Q + QQQQ Q Q Q
Sbjct: 635 LIGMLFPGDMVVFKPQIP-NQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQ 693
Query: 61 QHHHHSMKHGFPPFSSSKNKQQQQSSQMSDEDE 93
Q H + S ++ QQQQ Q + +
Sbjct: 694 QQQHQQQQQQQHQLSQLQHHQQQQQQQQQQQQQ 726
Score = 37.7 bits (86), Expect = 0.014
Identities = 27/90 (30%), Positives = 35/90 (38%), Gaps = 8/90 (8%)
Query: 19 LHQQQQQQKQQNLPNQQQQNPHQL----HHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPF 74
L QQQQ+QQ QQQQ HQL HH Q A + + M+ P
Sbjct: 710 LQHHQQQQQQQ----QQQQQQHQLTQLQHHHQQQQQASPLNQMQQQTSPLNQMQQQTSPL 765
Query: 75 SSSKNKQQQQSSQMSDEDEPNFPAEESSGG 104
+ + +QQ Q M + P GG
Sbjct: 766 NQMQQQQQPQQMVMGGQAFAQAPGRSQQGG 795
Score = 31.6 bits (70), Expect = 1.0
Identities = 18/85 (21%), Positives = 46/85 (53%), Gaps = 2/85 (2%)
Query: 406 KQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLENERM 465
K I N+ Q ++Q + Q Q+++Q+ + ++ ++++ + ++++++ + ++
Sbjct: 648 KPQIPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQL 707
Query: 466 VLLIRKKELELMHIQQQQQQQHSST 490
L ++ + QQQQQQQH T
Sbjct: 708 SQLQHHQQQQQQ--QQQQQQQHQLT 730
Score = 29.6 bits (65), Expect = 3.9
Identities = 13/20 (65%), Positives = 13/20 (65%)
Query: 268 PPQNQPQQQHQHQHQQQNQQ 287
P Q Q QQQ HQ QQQ QQ
Sbjct: 652 PNQQQQQQQQLHQQQQQQQQ 671
Score = 28.5 bits (62), Expect = 8.8
Identities = 12/19 (63%), Positives = 13/19 (68%)
Query: 268 PPQNQPQQQHQHQHQQQNQ 286
P Q QQQHQ Q QQQ+Q
Sbjct: 688 PQLQQQQQQHQQQQQQQHQ 706
>At4g32551 Leunig protein
Length = 931
Score = 42.7 bits (99), Expect = 5e-04
Identities = 30/107 (28%), Positives = 46/107 (42%), Gaps = 6/107 (5%)
Query: 17 IPLHQQQQQQKQ--QNLPNQQQQNPHQLHHSQ-VVSYAPQHHDTDTNQHHHHSMKHGFPP 73
I +QQ QQ Q Q QQQQ Q+ Q ++ A Q QHHHH +
Sbjct: 84 IKAREQQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQ---Q 140
Query: 74 FSSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKRKISPWQRMKWTD 120
+ +QQQQ Q P+ ++ S +++ +P Q+ + D
Sbjct: 141 QQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTPQQQPQRRD 187
Score = 33.1 bits (74), Expect = 0.36
Identities = 25/95 (26%), Positives = 36/95 (37%)
Query: 21 QQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNK 80
QQQQQQ+QQ QQQQ+ +Q Q + H + G + S N
Sbjct: 139 QQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTPQQQPQRRDGSHLANGSANG 198
Query: 81 QQQQSSQMSDEDEPNFPAEESSGGDPKRKISPWQR 115
+S+ P + +S +R P QR
Sbjct: 199 LVGNNSEPVMRQNPGSGSSLASKAYEERVKMPTQR 233
Score = 32.7 bits (73), Expect = 0.47
Identities = 26/97 (26%), Positives = 35/97 (35%), Gaps = 23/97 (23%)
Query: 21 QQQQQQKQQNLP--------------NQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHS 66
QQQQQQ+QQ + QQQQ+ H Q Q QH +
Sbjct: 101 QQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQ- 159
Query: 67 MKHGFPPFSSSKNKQQQQSSQMSDEDEPNFPAEESSG 103
PP S+ +QQQ + Q + P + G
Sbjct: 160 -----PP---SQQQQQQSTPQHQQQPTPQQQPQRRDG 188
Score = 30.4 bits (67), Expect = 2.3
Identities = 13/19 (68%), Positives = 13/19 (68%)
Query: 270 QNQPQQQHQHQHQQQNQQH 288
Q Q QQQ Q Q QQQ QQH
Sbjct: 138 QQQQQQQQQQQQQQQQQQH 156
Score = 30.0 bits (66), Expect = 3.0
Identities = 18/70 (25%), Positives = 30/70 (42%), Gaps = 1/70 (1%)
Query: 21 QQQQQQKQQNLPNQQQQNPHQLHHSQVVSYAPQHHDTDTNQHHHHSMKHGFPPFSSSKNK 80
QQQQQQ+ + QQQQ Q Q Q+ Q + +H P + +
Sbjct: 125 QQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQP-TPQQQP 183
Query: 81 QQQQSSQMSD 90
Q++ S +++
Sbjct: 184 QRRDGSHLAN 193
Score = 29.6 bits (65), Expect = 3.9
Identities = 12/18 (66%), Positives = 12/18 (66%)
Query: 270 QNQPQQQHQHQHQQQNQQ 287
Q Q QQQ H HQQQ QQ
Sbjct: 124 QQQQQQQQHHHHQQQQQQ 141
Score = 29.6 bits (65), Expect = 3.9
Identities = 12/18 (66%), Positives = 12/18 (66%)
Query: 270 QNQPQQQHQHQHQQQNQQ 287
Q Q QQQH H QQQ QQ
Sbjct: 125 QQQQQQQHHHHQQQQQQQ 142
Score = 29.6 bits (65), Expect = 3.9
Identities = 12/18 (66%), Positives = 12/18 (66%)
Query: 270 QNQPQQQHQHQHQQQNQQ 287
Q Q QQQ QH H QQ QQ
Sbjct: 123 QQQQQQQQQHHHHQQQQQ 140
Score = 29.6 bits (65), Expect = 3.9
Identities = 12/18 (66%), Positives = 12/18 (66%)
Query: 270 QNQPQQQHQHQHQQQNQQ 287
Q Q QQ H HQ QQQ QQ
Sbjct: 126 QQQQQQHHHHQQQQQQQQ 143
Score = 29.3 bits (64), Expect = 5.2
Identities = 17/105 (16%), Positives = 52/105 (49%), Gaps = 14/105 (13%)
Query: 385 NQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKK 444
N+ ++E++ + + ++Q +++ Q+ +Q+ + Q Q+++ L+ A+ ++
Sbjct: 68 NEKHSEVAASYIETQMIKAREQQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQ 127
Query: 445 EREMERAKLENERRRLENERMVLLIRKKELELMHIQQQQQQQHSS 489
+++ + ++++ + ++ QQQQQQQH +
Sbjct: 128 QQQQHHHHQQQQQQQQQQQQQ--------------QQQQQQQHQN 158
Score = 29.3 bits (64), Expect = 5.2
Identities = 14/63 (22%), Positives = 38/63 (60%)
Query: 424 ESQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLENERMVLLIRKKELELMHIQQQQ 483
E+Q + +Q+L+ +++ +++ ++ + + + ++L +R ++++ + H QQQQ
Sbjct: 80 ETQMIKAREQQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQ 139
Query: 484 QQQ 486
QQQ
Sbjct: 140 QQQ 142
Score = 28.9 bits (63), Expect = 6.7
Identities = 20/52 (38%), Positives = 25/52 (47%), Gaps = 10/52 (19%)
Query: 256 QHQVEGGSGTTTPPQNQPQQQHQHQ-----HQQQNQQHCFHSSE--NGVGSL 300
Q+Q +GG PPQ QPQ Q +Q Q Q+ H H E G GS+
Sbjct: 453 QNQQQGGGN---PPQPQPQPQPLNQLALTNPQPQSSNHSIHQQEKLGGGGSI 501
Score = 28.5 bits (62), Expect = 8.8
Identities = 56/262 (21%), Positives = 93/262 (35%), Gaps = 45/262 (17%)
Query: 256 QHQVEGGSGTTTPPQNQPQQQHQHQHQQQ----------NQQHCFHSSENGV---GSLGV 302
Q Q + PP Q QQQ QHQQQ + H + S NG+ S V
Sbjct: 148 QQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTPQQQPQRRDGSHLANGSANGLVGNNSEPV 207
Query: 303 SRGEGLRMLKIGSGYVEEEDDEDEEDESEDFS--DEGEDESGEGCSKGHINDQDEEENDG 360
R + S EE + ES D + D G+ H + G
Sbjct: 208 MRQNPGSGSSLASKAYEERVKMPTQRESLDEAAMKRFGDNVGQLLDPSHASILKSAAASG 267
Query: 361 KPSRK--RARKGGFSFPRSSSSTQLVN---QMNNEISGVF----------------QDGG 399
+P+ + + GG S + + QL + +EI+ V + G
Sbjct: 268 QPAGQVLHSTSGGMSPQVQTRNQQLPGSAVDIKSEINPVLTPRTAVPEGSLIGIPGSNQG 327
Query: 400 KSTWEKKQWIRNRIMQ-----LEEQKIGYESQAFQLEKQRLKWARYSSKKEREMERAKLE 454
+ K W Q L++QK +SQ+F +L +++ + + L
Sbjct: 328 SNNLTLKGWPLTGFDQLRSGLLQQQKPFMQSQSF----HQLNMLTPQHQQQLMLAQQNLN 383
Query: 455 NERRRLENERMVLLIRKKELEL 476
++ EN R+ +L+ + + L
Sbjct: 384 SQSVSEENRRLKMLLNNRSMTL 405
>At2g19480 putative nucleosome assembly protein
Length = 379
Score = 42.7 bits (99), Expect = 5e-04
Identities = 21/50 (42%), Positives = 31/50 (62%), Gaps = 1/50 (2%)
Query: 321 EDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKG 370
EDD+DE DE +D DE +DE E +D++EE + GK S+K++ G
Sbjct: 307 EDDDDEIDEDDDEEDEEDDEDDEE-EDDEDDDEEEEADQGKKSKKKSSAG 355
>At5g16030 unknown protein
Length = 339
Score = 41.6 bits (96), Expect = 0.001
Identities = 21/58 (36%), Positives = 34/58 (58%), Gaps = 11/58 (18%)
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRS 377
EED+E+EE+E +D S+E + E ++DE+E + K +K+ G FS+ RS
Sbjct: 270 EEDEEEEEEEKQDMSEEDDKE-----------EEDEQEEEEKTKKKKRGPGCFSWVRS 316
Score = 30.8 bits (68), Expect = 1.8
Identities = 17/58 (29%), Positives = 28/58 (47%), Gaps = 11/58 (18%)
Query: 318 VEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFP 375
+ EEDD++EEDE E+ + + G GC + +++ARK + FP
Sbjct: 283 MSEEDDKEEEDEQEEEEKTKKKKRGPGCFSW-----------VRSRQRQARKSKYIFP 329
>At3g06130 unknown protein
Length = 473
Score = 41.6 bits (96), Expect = 0.001
Identities = 43/181 (23%), Positives = 72/181 (39%), Gaps = 29/181 (16%)
Query: 235 FREMCAYHNSCGHGGVATSNVQHQVE---------GGSGTTTPPQNQPQQQHQHQHQQQN 285
F+ M H G GG +N + + GG G + P+ Q QH Q Q
Sbjct: 92 FKGMQIDHGKAGGGGGGNNNNNKKGQKNGGGGGGGGGGGNSNAPKMGQQLNPQHLQQLQQ 151
Query: 286 QQHCFHSSENGV-----GSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDESEDFSDEGEDE 340
Q + + GS+ V++ + + +K D E+D+ +DFSDE +DE
Sbjct: 152 LQKMKGFQDLKLPPQLKGSVPVNKNQNQKGVKF---------DVPEDDDDDDFSDEFDDE 202
Query: 341 SGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSSSSTQLVNQMN-NEISGVFQDGG 399
+ +D+ ++E D P K + ++ + Q N N G ++GG
Sbjct: 203 FTD-----DDDDEFDDEFDDLPLPSNKMKPNMTMMPNAQQMMMNAQKNANLAGGPAKNGG 257
Query: 400 K 400
K
Sbjct: 258 K 258
>At1g76890 unknown protein
Length = 575
Score = 40.8 bits (94), Expect = 0.002
Identities = 29/109 (26%), Positives = 50/109 (45%), Gaps = 26/109 (23%)
Query: 100 ESSGGDPKRKISPWQRMKWTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKSSGLLQKKGK- 158
ESSGG + + MK +T G+ AGS G + ++ LL+ + +
Sbjct: 10 ESSGGGVGGSVEEEKDMKMEET------------GEGAGSGGNRWPRPETLALLRIRSEM 57
Query: 159 -------------WKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVND 194
W+ +SR MME G+ S ++C++KF ++ K +KR +
Sbjct: 58 DKAFRDSTLKAPLWEEISRKMMELGYKRSSKKCKEKFENVYKYHKRTKE 106
Score = 38.5 bits (88), Expect = 0.008
Identities = 54/290 (18%), Positives = 96/290 (32%), Gaps = 80/290 (27%)
Query: 65 HSMKHGFPPFSSSKNKQQQQSSQMSDEDEPNFPAEES-----------SGGDPKRKISPW 113
H + G P N + Q Q + F ++E D +SP
Sbjct: 335 HKISGGQPQQPQQHNHKPSQRKQYQSDHSITFESKEPRAVLLDTTIKMGNYDNNHSVSP- 393
Query: 114 QRMKWTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKSSGLLQKKGKWKSVSRAMMEKGFYV 173
+W T V LI + GT K W+ +S M G+
Sbjct: 394 SSSRWPKTEVEALIRIRKNLEANYQENGT------------KGPLWEEISAGMRRLGYNR 441
Query: 174 SPQQCEDKFNDLNKRYKRVNDILGKGTACRVVENQGLLDSMDLSPKMKDEVRKLLNSKHL 233
S ++C++K+ ++NK +K+V K ++ R L +
Sbjct: 442 SAKRCKEKWENINKYFKKV--------------------------KESNKKRPLDSKTCP 475
Query: 234 FFREMCAYHNSCGHGGVATSNVQHQVEGGSGTTTPPQNQPQQQHQHQHQQQNQQHCFHSS 293
+F ++ A +N + G P PQ+Q + Q + F +
Sbjct: 476 YFHQLEALYN------------ERNKSGAMPLPLPLMVTPQRQLLLSQETQTE---FETD 520
Query: 294 ENGVGSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDESEDFSDEGEDESGE 343
+ K+G EEE + +E++ E+ EG++E+ E
Sbjct: 521 QRE---------------KVGDKEDEEEGESEEDEYDEEEEGEGDNETSE 555
>At5g56950 nucleosome assembly protein
Length = 374
Score = 40.4 bits (93), Expect = 0.002
Identities = 24/61 (39%), Positives = 33/61 (53%), Gaps = 9/61 (14%)
Query: 314 GSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRA---RKG 370
G + + DDED+ DE ED +E EDE E +D+DEEE K +K + +KG
Sbjct: 301 GEEFEIDNDDEDDIDEDEDEDEEDEDEDEEE------DDEDEEEEVSKTKKKPSVLHKKG 354
Query: 371 G 371
G
Sbjct: 355 G 355
>At4g02720 unknown protein
Length = 422
Score = 40.4 bits (93), Expect = 0.002
Identities = 34/136 (25%), Positives = 62/136 (45%), Gaps = 11/136 (8%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSS 378
+ ED ++ DE +D + D++ +G + +D E E+DG SRKR +S
Sbjct: 70 QNEDSDENADEIQDKNGGERDDNSKGKERKGKSDS-ESESDGLRSRKR---------KSK 119
Query: 379 SSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWA 438
SS + + S +G +S E++ R R ++K S++F+ ++ +
Sbjct: 120 SSRSKRRRKRSYDSDSESEGSESDSEEED-RRRRRKSSSKRKKSRSSRSFRKKRSHRRKT 178
Query: 439 RYSSKKEREMERAKLE 454
+YS E E +K E
Sbjct: 179 KYSDSDESSDEDSKAE 194
Score = 28.9 bits (63), Expect = 6.7
Identities = 22/97 (22%), Positives = 46/97 (46%), Gaps = 8/97 (8%)
Query: 339 DESGEGCSKGHIN--DQDEEENDGKPSRKRARKGGFSFPRSSSSTQLVNQMNNEISGVFQ 396
DES + SK I+ EEE+ S++R + S RS + ++ G +
Sbjct: 184 DESSDEDSKAEISASSSGEEEDTKSKSKRRKKSSDSSSKRSKGEK---TKSGSDSDGTEE 240
Query: 397 DGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQ 433
D S + + ++N ++L+E+++ + +L+K+
Sbjct: 241 D---SKMQVDETVKNTELELDEEELKKFKEMIELKKK 274
>At3g09360 putative transcription factor
Length = 600
Score = 40.4 bits (93), Expect = 0.002
Identities = 58/240 (24%), Positives = 98/240 (40%), Gaps = 46/240 (19%)
Query: 259 VEGGS-GTTTPPQNQPQQQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGY 317
V GG G + PP Q ++ + + + + +E G+ SL + E L L+I G
Sbjct: 319 VSGGLVGGSNPPAFQRAEKERMEKAAREE------NEGGISSL--NHDEQLYHLRIYLGC 370
Query: 318 VEEEDDEDEE---------DESEDFSDEGEDESGEGCSKGHINDQDE---------EEND 359
V E+ ++D++ DES++FSD +DE G+IN+++E E N
Sbjct: 371 VAEKGEKDKDGAEEHADTSDESDNFSDISDDE-----VNGYINNEEETHYKTITWTEMNK 425
Query: 360 G---KPSRKRARKGGFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQL 416
+ + K A S +S++ D KS EK+Q
Sbjct: 426 DYLEEQAAKEAALKAASEALKASNSNCPEDARKAFEAAKADAAKSRKEKQQKKAEEAKNA 485
Query: 417 EEQKIGYESQAFQLEKQRLKWA-RY--------SSKKEREMERAKLEN--ERRRLENERM 465
E+ L+K+RL Y +S E+ +R+K E E+++ EN+ M
Sbjct: 486 APPATAVEAVRRTLDKKRLSSVINYDVLESLFDTSAPEKSPKRSKTETDIEKKKEENKEM 545
Score = 28.5 bits (62), Expect = 8.8
Identities = 16/45 (35%), Positives = 30/45 (66%), Gaps = 4/45 (8%)
Query: 315 SGYVEEEDDEDEEDES-EDFSDEGEDESGEGCSKGHINDQDEEEN 358
+G E+ED+EDEE+ + E + + + ++GE K + D++EEE+
Sbjct: 552 NGENEDEDEEDEEEGNVESYDMKTDFQNGE---KFYEEDEEEEED 593
>At5g17690 chromo domain protein
Length = 445
Score = 40.0 bits (92), Expect = 0.003
Identities = 23/53 (43%), Positives = 32/53 (59%), Gaps = 2/53 (3%)
Query: 293 SENGVGSLGVSRGEGLRMLKIGSGY-VEEEDDEDEEDESEDFSDEGEDESGEG 344
S++G G G GE + + +IG E+ D+E+EEDE ED + EDE GEG
Sbjct: 42 SDDGGGGGGGGSGESI-LREIGDDRPTEDGDEEEEEDEDEDDGGDEEDEEGEG 93
>At3g25990 putative DNA-binding protein, GT-1
Length = 372
Score = 40.0 bits (92), Expect = 0.003
Identities = 32/104 (30%), Positives = 48/104 (45%), Gaps = 14/104 (13%)
Query: 100 ESSGGDPKRKI-SPWQRMK-WTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKSSGLLQKKG 157
ESSGG+ I +P +R + W R LI + + N KS+ K
Sbjct: 35 ESSGGEDHEIIKAPKKRAETWAQDETRTLISLRREMDNLF-------NTSKSN-----KH 82
Query: 158 KWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVNDILGKGTA 201
W+ +S+ M EKGF SP C DK+ ++ K +K+ K T+
Sbjct: 83 LWEQISKKMREKGFDRSPSMCTDKWRNILKEFKKAKQHEDKATS 126
>At1g56660 hypothetical protein
Length = 522
Score = 40.0 bits (92), Expect = 0.003
Identities = 44/219 (20%), Positives = 95/219 (43%), Gaps = 20/219 (9%)
Query: 275 QQHQHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDESEDFS 334
++H+ +H++ ++ E G ++ E K SG E+ D+E + ED S
Sbjct: 109 EEHEKEHKKGKEKKHEELEEEKEGKKKKNKKE-----KDESGPEEKNKKADKEKKHEDVS 163
Query: 335 DEGEDESGEGCSKGHINDQDE---EENDGKPSRKRARKGGFSFPRSSSSTQLVNQMNNEI 391
E E+ E K ++DE EE KP +++ +K S S + + ++
Sbjct: 164 QEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKKQK------EESKSNE-----DKKV 212
Query: 392 SGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKEREMERA 451
G + G K EK+ + + +Q++ + +K++ + KK+ + E+
Sbjct: 213 KGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEK- 271
Query: 452 KLENERRRLENERMVLLIRKKELELMHIQQQQQQQHSST 490
K ++E E++++ K E + ++ ++H +T
Sbjct: 272 KEKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTKEHDAT 310
Score = 32.7 bits (73), Expect = 0.47
Identities = 31/145 (21%), Positives = 67/145 (45%), Gaps = 10/145 (6%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSS 378
EE + E ES+ +E E E +G K H ++E+E K ++K + G P
Sbjct: 93 EEGHGDLEVKESDVKVEEHEKEHKKGKEKKHEELEEEKEGKKKKNKKEKDESG---PEEK 149
Query: 379 SSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWA 438
+ + + ++S K E++ +N+ + E+ + G E + + +K++ +
Sbjct: 150 NKKADKEKKHEDVS-----QEKEELEEEDGKKNK--KKEKDESGTEEKKKKPKKEKKQKE 202
Query: 439 RYSSKKEREMERAKLENERRRLENE 463
S ++++++ K + E+ LE E
Sbjct: 203 ESKSNEDKKVKGKKEKGEKGDLEKE 227
Score = 31.2 bits (69), Expect = 1.4
Identities = 81/452 (17%), Positives = 167/452 (36%), Gaps = 65/452 (14%)
Query: 54 HHDTDTNQHHHHSMKHGFPPFSSSKNKQQQQSSQMSDEDEPNFPAEESSGGDPKRKISPW 113
H D + + +H + K ++ + + + N ++ SG + K K +
Sbjct: 96 HGDLEVKESDVKVEEHEKEHKKGKEKKHEELEEEKEGKKKKNKKEKDESGPEEKNKKADK 155
Query: 114 QRMKWTDTMVRLLIMAVYYIGDEAGSEGTDPNKKKS---SGLLQKKGKWKSVSRAMMEKG 170
++ K D V +E E NKKK SG +KK K K +
Sbjct: 156 EK-KHED---------VSQEKEELEEEDGKKNKKKEKDESGTEEKKKKPKKEKK------ 199
Query: 171 FYVSPQQCEDKFNDLNKRYKRVNDILGKGTACRVVENQGLL-DSMDLSPKMKDEVRKLLN 229
Q+ E K N+ +K+ K + KG + E + D D K KD +
Sbjct: 200 -----QKEESKSNE-DKKVKGKKEKGEKGDLEKEDEEKKKEHDETDQEMKEKDSKKNKKK 253
Query: 230 SKHLFFREMCAYHNSCG------HGGVATSNVQHQVEGGSGTTTPPQNQPQQQHQHQHQQ 283
K E CA +T +++G G P+ + + + +H
Sbjct: 254 EKD----ESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPEKEDEGKKTKEHDA 309
Query: 284 QNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDDEDEEDE----------SEDF 333
Q+ ++++ G ++ + + + E+E + ++DE E
Sbjct: 310 TEQEMDDEAADHKEGKKKKNKDKAKKKETVIDEVCEKETKDKDDDEGETKQKKNKKKEKK 369
Query: 334 SDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSSSSTQLVNQMNNEISG 393
S++GE + E K + + + D K A K + T+ + +++ G
Sbjct: 370 SEKGEKDVKEDKKKENPLETEVMSRDIKLEEPEAEK------KEEDDTE--EKKKSKVEG 421
Query: 394 VFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSKKERE------ 447
+ GK +KK +N+ +E K+ + + + + + +K +K+E++
Sbjct: 422 GESEEGKKK-KKKDKKKNKKKDTKEPKMTEDEEEKKDDSKDVKIEGSKAKEEKKDKDVKK 480
Query: 448 ----MERAKLENERRRLENERMVLLIRKKELE 475
+ KL+ + +++ + L+ K E+E
Sbjct: 481 KKGGNDIGKLKTKLAKIDEKIGALMEEKAEIE 512
Score = 28.5 bits (62), Expect = 8.8
Identities = 32/162 (19%), Positives = 66/162 (39%), Gaps = 18/162 (11%)
Query: 304 RGEGLRM-LKIGSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEE-ENDGK 361
+GE + + +++ + +E+ + +E+ S + E + G+ D+D++ + DGK
Sbjct: 27 KGENVEVEMEVKAKSIEKVKAKKDEESSGKSKKDKEKKKGKNVDSEVKEDKDDDKKKDGK 86
Query: 362 PSRKRARKGGFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKI 421
K+ +G S ++ G EKK +LEE+K
Sbjct: 87 MVSKKHEEGHGDLEVKESDVKVEEHEKEHKKGK---------EKKH------EELEEEKE 131
Query: 422 GYESQAFQLEKQRLKWARYSSKKEREMERAKLENERRRLENE 463
G + + + EK + K ++E + + E+ LE E
Sbjct: 132 G-KKKKNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELEEE 172
>At5g53800 unknown protein
Length = 339
Score = 39.7 bits (91), Expect = 0.004
Identities = 35/150 (23%), Positives = 68/150 (45%), Gaps = 17/150 (11%)
Query: 320 EEDDEDEEDESEDFSDEGEDESGE-GCSKGHINDQDEE----------ENDGKPSRKRAR 368
EE D ES+ + +G D E G G ++++E ++D K SR R R
Sbjct: 40 EEKDVSRRRESKRRTKDGNDSGSESGLESGSESEKEERRRSRKDRGKRKSDRKSSRSRRR 99
Query: 369 KGGFSFPRSSSSTQLVNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAF 428
+ +S S S ++ ++ ++ +D E+++ R R + EE+K +
Sbjct: 100 RRDYSSSSSDSESESESEYSDSEESESED------ERRRRKRKRKEREEEEKERKRRRRE 153
Query: 429 QLEKQRLKWARYSSKKEREMERAKLENERR 458
+ +K+R K + KK +E ++ K E ++
Sbjct: 154 KDKKKRNKSDKDGDKKRKEKKKKKSEKVKK 183
>At5g03710 putative protein
Length = 81
Score = 39.7 bits (91), Expect = 0.004
Identities = 19/56 (33%), Positives = 35/56 (61%), Gaps = 1/56 (1%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSF 374
EEE++E+EE+E E+ +E E+E E + +++EEE + + R+R + F+F
Sbjct: 23 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDREREER-AFAF 77
Score = 38.1 bits (87), Expect = 0.011
Identities = 17/51 (33%), Positives = 31/51 (60%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARK 369
EEE++E+EE+E E+ +E E+E E + +++EEE + + R R+
Sbjct: 21 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDRERE 71
Score = 36.6 bits (83), Expect = 0.032
Identities = 16/51 (31%), Positives = 31/51 (60%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARK 369
EEE++E+EE+E E+ +E E+E E + +++EEE + + + R+
Sbjct: 19 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDRE 69
Score = 36.2 bits (82), Expect = 0.042
Identities = 16/44 (36%), Positives = 28/44 (63%)
Query: 316 GYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
G EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 5 GEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 48
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 14 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 54
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 12 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 52
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 15 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 55
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 11 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 51
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 18 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 58
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 16 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 56
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 17 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 57
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 10 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 50
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 9 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 49
Score = 35.4 bits (80), Expect = 0.072
Identities = 15/41 (36%), Positives = 27/41 (65%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEEND 359
EEE++E+EE+E E+ +E E+E E + +++EEE +
Sbjct: 13 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 53
>At3g51070 putative protein
Length = 895
Score = 39.7 bits (91), Expect = 0.004
Identities = 34/136 (25%), Positives = 57/136 (41%), Gaps = 27/136 (19%)
Query: 256 QHQVEGGSGTTTPPQN-----QPQQQHQHQHQ--QQNQQHCFHSSENGVGSLGVSRGEGL 308
+ +E G G T+ QP++Q+ + QQN++ S ENG G
Sbjct: 221 EQPMETGQGETSETSKNEENGQPEEQNSGNEETGQQNEEKTTASEENGKGE--------- 271
Query: 309 RMLKIGSGYVEEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRAR 368
+ +K +G EE +EE +++ +DE+ E Q EE D K + +
Sbjct: 272 KSMKDENGQQEEHTTAEEESGNKEEESTSKDENME---------QQEERKDEKKHEQGSE 322
Query: 369 KGGF--SFPRSSSSTQ 382
GF P+ S+ +Q
Sbjct: 323 ASGFGSGIPKESAESQ 338
Score = 36.2 bits (82), Expect = 0.042
Identities = 45/247 (18%), Positives = 92/247 (37%), Gaps = 22/247 (8%)
Query: 206 ENQGLLDSMDLSPKMKDEVRKLLNSKHLFFREMCAYHNSCGHGGVATSNVQHQVEGGSGT 265
+N G L D + K +DE RK K + + S + + G
Sbjct: 89 DNPGKLP--DDAVKSEDEQRKSAKEKSETTSSKTQTQETQQNNDDKISEEKEKDNGKENQ 146
Query: 266 TTPPQNQPQQQH--QHQHQQQNQQHCFHSSENGVGSLGVSRGEGLRMLKIGSGYVEEEDD 323
T + Q + + ++Q QQ + G+ G +G+G + G + ++
Sbjct: 147 TVQESEEGQMKKVVKEFEKEQKQQRDEDAGTQPKGTQGQEQGQGKEQPDVEQGNKQGQEQ 206
Query: 324 EDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKRARKGGFSFPRSSSSTQL 383
+ D + F+D + E +G ++ + E +G+P + +S +
Sbjct: 207 DSNTDVT--FTDATKQEQPMETGQGETSETSKNEENGQPEEQ------------NSGNEE 252
Query: 384 VNQMNNEISGVFQDGGKSTWEKKQWIRNRIMQLEEQKIGYESQAFQLEKQRLKWARYSSK 443
Q N E + ++ GK ++ +++ Q EE E + E+ K +
Sbjct: 253 TGQQNEEKTTASEENGKG----EKSMKDENGQQEEHTTAEEESGNKEEESTSKDENMEQQ 308
Query: 444 KEREMER 450
+ER+ E+
Sbjct: 309 EERKDEK 315
>At2g22080 En/Spm-like transposon protein
Length = 140
Score = 39.7 bits (91), Expect = 0.004
Identities = 16/48 (33%), Positives = 31/48 (64%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGKPSRKR 366
++ED+E++EDE++D +E +DE E + D+D+EE P +++
Sbjct: 92 DDEDEEEDEDENDDGGEEDDDEDAEVEEEEEEEDEDDEEALQPPKKRK 139
Score = 33.1 bits (74), Expect = 0.36
Identities = 13/42 (30%), Positives = 25/42 (58%)
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEEENDGK 361
E D D+E E +D +D+ +D++ E + D+DE ++ G+
Sbjct: 67 EAGDNDDEPEGDDGNDDEDDDNHENDDEDEEEDEDENDDGGE 108
Score = 32.7 bits (73), Expect = 0.47
Identities = 17/60 (28%), Positives = 36/60 (59%), Gaps = 11/60 (18%)
Query: 319 EEEDDEDEEDESEDFSDEGEDESGEGCSK-----GHINDQDEEENDG-----KPSRKRAR 368
++EDD++ E++ ED +E EDE+ +G + + +++EEE++ +P +KR +
Sbjct: 82 DDEDDDNHENDDED-EEEDEDENDDGGEEDDDEDAEVEEEEEEEDEDDEEALQPPKKRKK 140
Score = 28.9 bits (63), Expect = 6.7
Identities = 11/37 (29%), Positives = 22/37 (58%)
Query: 320 EEDDEDEEDESEDFSDEGEDESGEGCSKGHINDQDEE 356
++ ++DE+D++ + DE E+E + G D DE+
Sbjct: 78 DDGNDDEDDDNHENDDEDEEEDEDENDDGGEEDDDED 114
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.128 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,720,273
Number of Sequences: 26719
Number of extensions: 658573
Number of successful extensions: 11140
Number of sequences better than 10.0: 581
Number of HSP's better than 10.0 without gapping: 286
Number of HSP's successfully gapped in prelim test: 307
Number of HSP's that attempted gapping in prelim test: 5519
Number of HSP's gapped (non-prelim): 2752
length of query: 490
length of database: 11,318,596
effective HSP length: 103
effective length of query: 387
effective length of database: 8,566,539
effective search space: 3315250593
effective search space used: 3315250593
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139525.4


