

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.10 + phase: 0
(125 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
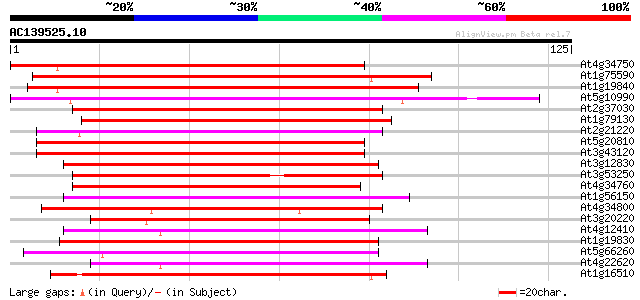
Score E
Sequences producing significant alignments: (bits) Value
At4g34750 putative protein 91 1e-19
At1g75590 unknown protein 89 4e-19
At1g19840 hypothetical protein 86 5e-18
At5g10990 unknown protein 83 3e-17
At2g37030 putative auxin-induced protein 75 8e-15
At1g79130 unknown protein 72 5e-14
At2g21220 auxin-regulated like protein 70 4e-13
At5g20810 unknown protein 69 6e-13
At3g43120 unknown protein 69 6e-13
At3g12830 unknown protein 68 1e-12
At3g53250 putative protein 68 1e-12
At4g34760 putative auxin-regulated protein 67 2e-12
At1g56150 unknown protein 67 2e-12
At4g34800 putative protein 66 5e-12
At3g20220 unknown protein 65 1e-11
At4g12410 putative protein 64 1e-11
At1g19830 unknown protein 64 2e-11
At5g66260 auxin-induced protein-like 63 3e-11
At4g22620 putative protein 63 3e-11
At1g16510 auxin-induced like protein 63 4e-11
>At4g34750 putative protein
Length = 150
Score = 90.9 bits (224), Expect = 1e-19
Identities = 45/82 (54%), Positives = 56/82 (67%), Gaps = 3/82 (3%)
Query: 1 MACMWRKNA---CSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEG 57
M W+K A S SDVP GH+AV+VGE RR+V+RA +LNHP+ + LL +A E
Sbjct: 16 MLKQWQKKAHIGSSNNDPVSDVPPGHVAVSVGENRRRYVVRAKHLNHPIFRRLLAEAEEE 75
Query: 58 YGFNKSGPLSIPCDEFLFEDIL 79
YGF GPL+IPCDE LFEDI+
Sbjct: 76 YGFANVGPLAIPCDESLFEDII 97
>At1g75590 unknown protein
Length = 154
Score = 89.4 bits (220), Expect = 4e-19
Identities = 46/91 (50%), Positives = 62/91 (67%), Gaps = 2/91 (2%)
Query: 6 RKNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGP 65
R ++ + +PSDVP GH+AV VG + RRFV+RA YLNHPVL+ LL QA E +GF GP
Sbjct: 29 RMSSSFSRCVPSDVPSGHVAVYVGSSCRRFVVRATYLNHPVLRNLLVQAEEEFGFVNQGP 88
Query: 66 LSIPCDEFLFEDIL--LSLGGGTVARRSSSP 94
L IPC+E +FE+ + +S T +RR + P
Sbjct: 89 LVIPCEESVFEESIRFISRSDSTRSRRFTCP 119
>At1g19840 hypothetical protein
Length = 153
Score = 85.9 bits (211), Expect = 5e-18
Identities = 43/90 (47%), Positives = 56/90 (61%), Gaps = 3/90 (3%)
Query: 5 WRKNA---CSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFN 61
WR A + +PSDVP GH+AV VG RRFV+RA YLNHP++ LL QA E +GF
Sbjct: 24 WRNKARLSSVSRCVPSDVPSGHVAVCVGSGCRRFVVRASYLNHPIISNLLVQAEEEFGFA 83
Query: 62 KSGPLSIPCDEFLFEDILLSLGGGTVARRS 91
GPL IPC+E +FE+ + + +R S
Sbjct: 84 NQGPLVIPCEESVFEEAIRFISRSDSSRSS 113
>At5g10990 unknown protein
Length = 148
Score = 83.2 bits (204), Expect = 3e-17
Identities = 50/127 (39%), Positives = 70/127 (54%), Gaps = 11/127 (8%)
Query: 1 MACMWRKNACSG---KKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEG 57
M WR A + +PSDVP GH+AV VG + RRFV+ A YLNHP+L LL +A E
Sbjct: 20 MLRQWRNKARMSSVRRSVPSDVPSGHVAVYVGRSCRRFVVLATYLNHPILMNLLVKAEEE 79
Query: 58 YGFNKSGPLSIPCDEFLFEDILLSLGGGT------VARRSSSPVLTKKLDLSFLKDAVPL 111
+GF GPL IPC+E +FE+ + + + +++ + KLDL L ++ PL
Sbjct: 80 FGFANQGPLVIPCEESVFEESIRFITRSSRFTCTDDLKKNRHGGIRSKLDL--LMESRPL 137
Query: 112 LEAFDSK 118
L K
Sbjct: 138 LHGVSEK 144
>At2g37030 putative auxin-induced protein
Length = 124
Score = 75.1 bits (183), Expect = 8e-15
Identities = 35/69 (50%), Positives = 45/69 (64%)
Query: 15 LPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFL 74
+P DVP+GHL V VGE +RFVI + L HP+ Q LLDQA + YGF+ L IPC+E
Sbjct: 45 IPRDVPKGHLVVYVGEEYKRFVININLLKHPLFQALLDQAQDAYGFSADSRLWIPCNEST 104
Query: 75 FEDILLSLG 83
F D++ G
Sbjct: 105 FLDVVRCAG 113
>At1g79130 unknown protein
Length = 134
Score = 72.4 bits (176), Expect = 5e-14
Identities = 33/69 (47%), Positives = 46/69 (65%)
Query: 17 SDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLFE 76
S VP GH+ V VGE RFV+ A+ LNHPV LL+++ + YG+ + G L IPC+ F+FE
Sbjct: 46 SSVPSGHVPVNVGEDKERFVVSAELLNHPVFVGLLNRSAQEYGYTQKGVLHIPCNVFVFE 105
Query: 77 DILLSLGGG 85
++ SL G
Sbjct: 106 QVVESLRSG 114
>At2g21220 auxin-regulated like protein
Length = 104
Score = 69.7 bits (169), Expect = 4e-13
Identities = 33/78 (42%), Positives = 46/78 (58%), Gaps = 1/78 (1%)
Query: 7 KNACSGKK-LPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGP 65
KN C + LP DVP+GH V VGE R+++ +L HP + LL QA E +GFN
Sbjct: 27 KNQCYDEDGLPVDVPKGHFPVYVGEKRSRYIVPISFLTHPKFKSLLQQAEEEFGFNHDMG 86
Query: 66 LSIPCDEFLFEDILLSLG 83
L+IPC+E +F + +G
Sbjct: 87 LTIPCEEVVFRSLTSMIG 104
>At5g20810 unknown protein
Length = 165
Score = 68.9 bits (167), Expect = 6e-13
Identities = 32/73 (43%), Positives = 49/73 (66%)
Query: 7 KNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPL 66
+ C + P DVP+G+LAV VG RRF+I YL+H + + LL++A E +GF++SG L
Sbjct: 67 EETCQSPEPPHDVPKGNLAVYVGPELRRFIIPTSYLSHSLFKVLLEKAEEEFGFDQSGAL 126
Query: 67 SIPCDEFLFEDIL 79
+IPC+ F+ +L
Sbjct: 127 TIPCEVETFKYLL 139
>At3g43120 unknown protein
Length = 160
Score = 68.9 bits (167), Expect = 6e-13
Identities = 32/73 (43%), Positives = 49/73 (66%)
Query: 7 KNACSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPL 66
+ C + P DVP+G+LAV VG RRF+I ++L+H + + LL++A E YGF+ SG L
Sbjct: 67 ETTCQSPEPPPDVPKGYLAVYVGPELRRFIIPTNFLSHSLFKVLLEKAEEEYGFDHSGAL 126
Query: 67 SIPCDEFLFEDIL 79
+IPC+ F+ +L
Sbjct: 127 TIPCEVETFKYLL 139
>At3g12830 unknown protein
Length = 132
Score = 68.2 bits (165), Expect = 1e-12
Identities = 33/70 (47%), Positives = 45/70 (64%)
Query: 13 KKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDE 72
KK S VP GH+ V VG+ RFV+ A+ LNHPV LL+++ + YG+ + G L IPC
Sbjct: 44 KKQTSSVPEGHVPVYVGDEMERFVVSAELLNHPVFIGLLNRSAQEYGYEQKGVLQIPCHV 103
Query: 73 FLFEDILLSL 82
+FE I+ SL
Sbjct: 104 LVFERIMESL 113
>At3g53250 putative protein
Length = 109
Score = 67.8 bits (164), Expect = 1e-12
Identities = 34/69 (49%), Positives = 43/69 (62%), Gaps = 3/69 (4%)
Query: 15 LPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFL 74
+P DVPRGHL V VG+ +RFVI+ L HP+ + LLDQA + Y S L IPCDE
Sbjct: 33 IPKDVPRGHLVVYVGDDYKRFVIKMSLLTHPIFKALLDQAQDAY---NSSRLWIPCDENT 89
Query: 75 FEDILLSLG 83
F D++ G
Sbjct: 90 FLDVVRCSG 98
>At4g34760 putative auxin-regulated protein
Length = 107
Score = 67.4 bits (163), Expect = 2e-12
Identities = 29/64 (45%), Positives = 41/64 (63%)
Query: 15 LPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFL 74
LP DVP+GH V VGE R+++ +L HP Q LL +A E +GF+ L+IPCDE +
Sbjct: 39 LPLDVPKGHFPVYVGENRSRYIVPISFLTHPEFQSLLQRAEEEFGFDHDMGLTIPCDELV 98
Query: 75 FEDI 78
F+ +
Sbjct: 99 FQTL 102
>At1g56150 unknown protein
Length = 110
Score = 67.0 bits (162), Expect = 2e-12
Identities = 36/77 (46%), Positives = 45/77 (57%)
Query: 13 KKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDE 72
+K S VP GH+ V VG RFV+ A+ LNHPV LL Q+ + YG+ + G L IPC
Sbjct: 32 EKHKSWVPEGHVPVYVGHEMERFVVNAELLNHPVFVALLKQSAQEYGYEQQGVLRIPCHV 91
Query: 73 FLFEDILLSLGGGTVAR 89
+FE IL SL G R
Sbjct: 92 LVFERILESLRLGLADR 108
>At4g34800 putative protein
Length = 94
Score = 65.9 bits (159), Expect = 5e-12
Identities = 38/80 (47%), Positives = 49/80 (60%), Gaps = 4/80 (5%)
Query: 8 NACSGKKLPSDVPRGHLAVTVGE---TNRRFVIRADYLNHPVLQELLDQAYEGYGFNKS- 63
N+ +K S VP+GH+AV VGE + +RFV+ YLNHP Q LL +A E +GFN
Sbjct: 10 NSKQSQKQQSRVPKGHVAVYVGEEMESKKRFVVPISYLNHPSFQGLLSRAEEEFGFNHPI 69
Query: 64 GPLSIPCDEFLFEDILLSLG 83
G L+IPC E F +L S G
Sbjct: 70 GGLTIPCREETFVGLLNSYG 89
>At3g20220 unknown protein
Length = 118
Score = 64.7 bits (156), Expect = 1e-11
Identities = 30/63 (47%), Positives = 44/63 (69%), Gaps = 1/63 (1%)
Query: 19 VPRGHLAVTVG-ETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLFED 77
VPRGHLAV VG E +RFVI YL +P + L+D+ + +G++ G + IPC+E +FE+
Sbjct: 47 VPRGHLAVYVGREERQRFVIPTKYLQYPEFRSLMDEVADEFGYDHEGGIHIPCEESVFEE 106
Query: 78 ILL 80
IL+
Sbjct: 107 ILI 109
>At4g12410 putative protein
Length = 157
Score = 64.3 bits (155), Expect = 1e-11
Identities = 36/84 (42%), Positives = 49/84 (57%), Gaps = 3/84 (3%)
Query: 13 KKLPSDVPRGHLAVTVGETN---RRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIP 69
KK VPRGHL V VGE++ RR V+ Y NHP+ ELL+QA +GF++ G ++IP
Sbjct: 70 KKSNRVVPRGHLVVHVGESDDDTRRVVVPVIYFNHPLFGELLEQAERVHGFDQPGRITIP 129
Query: 70 CDEFLFEDILLSLGGGTVARRSSS 93
C FE + L + RR +S
Sbjct: 130 CRVSDFEKVQLRIAAWDHCRRKNS 153
>At1g19830 unknown protein
Length = 117
Score = 63.9 bits (154), Expect = 2e-11
Identities = 30/71 (42%), Positives = 43/71 (60%)
Query: 12 GKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCD 71
G LP DVP+GH V VG R+V+ +L P Q LL QA E +GF+ + L+IPC+
Sbjct: 41 GDSLPLDVPKGHFVVYVGGNRVRYVLPISFLTRPEFQLLLQQAEEEFGFDHNMGLTIPCE 100
Query: 72 EFLFEDILLSL 82
E F+ ++ S+
Sbjct: 101 EVAFKSLITSM 111
>At5g66260 auxin-induced protein-like
Length = 99
Score = 63.2 bits (152), Expect = 3e-11
Identities = 33/84 (39%), Positives = 47/84 (55%), Gaps = 5/84 (5%)
Query: 4 MWRKNACSGKKLPSDV-----PRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGY 58
M ++ + GKK DV P+GH V VG + R VI +L HP+ Q LL Q+ E +
Sbjct: 14 MLKRCSSLGKKSSVDVNFNGVPKGHFVVYVGHSRSRHVIPISFLTHPIFQMLLQQSEEEF 73
Query: 59 GFNKSGPLSIPCDEFLFEDILLSL 82
GF + L+IPCDE F ++ S+
Sbjct: 74 GFFQDNGLTIPCDEHFFRALISSI 97
>At4g22620 putative protein
Length = 160
Score = 63.2 bits (152), Expect = 3e-11
Identities = 34/78 (43%), Positives = 44/78 (55%), Gaps = 3/78 (3%)
Query: 19 VPRGHLAVTVGETN---RRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIPCDEFLF 75
VPRGHL V VGE+ RR V+ Y NHP+ ELL+QA YGF + G + IPC F
Sbjct: 79 VPRGHLVVHVGESGEDTRRVVVPVIYFNHPLFGELLEQAERVYGFEQPGRIMIPCRVSDF 138
Query: 76 EDILLSLGGGTVARRSSS 93
E + + + RR S+
Sbjct: 139 EKVQMRIAAWDHCRRKST 156
>At1g16510 auxin-induced like protein
Length = 147
Score = 62.8 bits (151), Expect = 4e-11
Identities = 31/77 (40%), Positives = 49/77 (63%), Gaps = 3/77 (3%)
Query: 10 CSGKKLPSDVPRGHLAVTVGETNRRFVIRADYLNHPVLQELLDQAYEGYGFNKSGPLSIP 69
C +++ + VP GH+ V VGE RFV+ A+ +NHP+ LL+++ + YG+ + G L IP
Sbjct: 46 CPARRV-NTVPAGHVPVYVGEEMERFVVSAELMNHPIFVGLLNRSAQEYGYAQKGVLHIP 104
Query: 70 CDEFLFEDIL--LSLGG 84
C +FE ++ L LGG
Sbjct: 105 CHVIVFERVVETLRLGG 121
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,995,379
Number of Sequences: 26719
Number of extensions: 120643
Number of successful extensions: 365
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 247
Number of HSP's gapped (non-prelim): 87
length of query: 125
length of database: 11,318,596
effective HSP length: 87
effective length of query: 38
effective length of database: 8,994,043
effective search space: 341773634
effective search space used: 341773634
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC139525.10


