

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139525.1 - phase: 0
(395 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
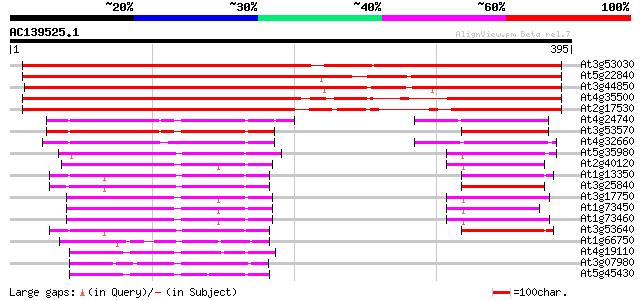
Score E
Sequences producing significant alignments: (bits) Value
At3g53030 serine protein kinase - like 572 e-163
At5g22840 serine protein kinase-like protein 558 e-159
At3g44850 putative protein 548 e-156
At4g35500 protein kinase - like protein 356 1e-98
At2g17530 putative protein kinase 347 5e-96
At4g24740 protein kinase (AFC2) 113 2e-25
At3g53570 protein kinase (AME2/AFC1) 110 1e-24
At4g32660 protein kinase (AME3) 107 1e-23
At5g35980 protein kinase-like 103 2e-22
At2g40120 protein kinase like protein 77 1e-14
At1g13350 Unknown protein 74 1e-13
At3g25840 protein kinase like protein 72 5e-13
At3g17750 protein kinase, putative 72 7e-13
At1g73450 69 3e-12
At1g73460 69 3e-12
At3g53640 protein kinase -like protein 67 1e-11
At1g66750 unknown protein 57 1e-08
At4g19110 putative serine/threonine-protein kinase 56 4e-08
At3g07980 putative MAP3K epsilon protein kinase 55 7e-08
At5g45430 serine/threonine-protein kinase Mak (male germ cell-as... 54 1e-07
>At3g53030 serine protein kinase - like
Length = 529
Score = 572 bits (1473), Expect = e-163
Identities = 277/379 (73%), Positives = 314/379 (82%), Gaps = 10/379 (2%)
Query: 10 TESSDFTSEDEGTEDYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRY 69
++ ++TSEDEGTEDYRRGGYHAVRIGD+F +GRYVVQSKLGWGHFSTVWL+WD+ SRY
Sbjct: 8 SDGGEYTSEDEGTEDYRRGGYHAVRIGDSFKTGRYVVQSKLGWGHFSTVWLSWDTQSSRY 67
Query: 70 VALKVQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEY 129
VALKVQKSAQHYTEAA+DEITILQQIAEGDTDD KCVVKLLDHFKHSGPNGQHVCMVFEY
Sbjct: 68 VALKVQKSAQHYTEAAMDEITILQQIAEGDTDDTKCVVKLLDHFKHSGPNGQHVCMVFEY 127
Query: 130 LGDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLSTIDP 189
LGDNLLTLIKYSDYRG+PI MVKEIC+H+LVGLDYLHKQLSIIHTDLKPEN+LL STIDP
Sbjct: 128 LGDNLLTLIKYSDYRGLPIPMVKEICYHMLVGLDYLHKQLSIIHTDLKPENVLLPSTIDP 187
Query: 190 SKDPRKSGAPLILPNSKDKTMLESTAARDTKTSNGDFIKNHKKNIKRKAKQAAHGCAEKE 249
SKDPRKSGAPL+LP KD T+++ SNGDF+KN K RKAK +A G AE +
Sbjct: 188 SKDPRKSGAPLVLPTDKDNTVVD---------SNGDFVKNQKTGSHRKAKLSAQGHAENK 238
Query: 250 ASEGVDGNHETSGAVESSPNASSAREQASSSAGTSRLSDADGMKLKEQGNRRGSRTVRQK 309
+ D G+ + ++ + TS + DG + +QG ++GSR+ R+
Sbjct: 239 GNTESD-KVRGVGSPVNGKQCAAEKSVEEDCPSTSDAIELDGSEKGKQGGKKGSRSSRRH 297
Query: 310 LLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFEL 369
L+ASAD+KCKLVDFGNACWTYKQFT+DIQTRQYRCPEVILGSKYSTSADLWSFACICFEL
Sbjct: 298 LVASADLKCKLVDFGNACWTYKQFTSDIQTRQYRCPEVILGSKYSTSADLWSFACICFEL 357
Query: 370 ATGDVLFDPHSGDNFDRDE 388
TGDVLFDPHSGDN+DRDE
Sbjct: 358 VTGDVLFDPHSGDNYDRDE 376
>At5g22840 serine protein kinase-like protein
Length = 538
Score = 558 bits (1438), Expect = e-159
Identities = 266/382 (69%), Positives = 315/382 (81%), Gaps = 15/382 (3%)
Query: 10 TESSDFTSEDEGTEDYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRY 69
+++SD++SEDEGTEDYRRGGYH VR+GDTF +G YV+QSKLGWGHFSTVWLAWD+ +SRY
Sbjct: 11 SDASDYSSEDEGTEDYRRGGYHTVRVGDTFKNGSYVIQSKLGWGHFSTVWLAWDTLNSRY 70
Query: 70 VALKVQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEY 129
VALK+QKSAQHYTEAA+DEI IL+QIAEGD +DKKCVVKLLDHFKH+GPNGQHVCMVFEY
Sbjct: 71 VALKIQKSAQHYTEAAMDEIKILKQIAEGDAEDKKCVVKLLDHFKHAGPNGQHVCMVFEY 130
Query: 130 LGDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLSTIDP 189
LGDNLL++IKYSDYRG+P++MVKEICFHILVGLDYLH++LSIIHTD+KPENILL STIDP
Sbjct: 131 LGDNLLSVIKYSDYRGVPLHMVKEICFHILVGLDYLHRELSIIHTDIKPENILLCSTIDP 190
Query: 190 SKDPRKSGAPLILPNSKDKTMLESTAARD---TKTSNGDFIKNHKKNIKRKAKQAAHGCA 246
D RKSG PL+LP KDK + E ++ + T + D KN KK I++KAK+
Sbjct: 191 EADARKSGIPLVLPTVKDKAVPERPVEKEKPKSYTYSADLTKNQKKKIRKKAKKV----- 245
Query: 247 EKEASEGVDGNHETSGAVESSPNASSAREQASSSAGTSRLSDADGMKLKEQGNRRGSRTV 306
EG + N S E+ PN ++ E+ S + R+ DA+ + K +GNRRGS++
Sbjct: 246 -----EGSEENERDSSNSEARPNGNATVERLEES--SERVKDAENVSQKSRGNRRGSQST 298
Query: 307 RQKLLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACIC 366
RQKLLA D KCKLVDFGNACWTYKQFT+DIQTRQYRCPEV+LGSKYSTSAD+WSFACIC
Sbjct: 299 RQKLLADVDRKCKLVDFGNACWTYKQFTSDIQTRQYRCPEVVLGSKYSTSADMWSFACIC 358
Query: 367 FELATGDVLFDPHSGDNFDRDE 388
FELATGDVLFDPHSG+NF+RDE
Sbjct: 359 FELATGDVLFDPHSGENFERDE 380
>At3g44850 putative protein
Length = 534
Score = 548 bits (1412), Expect = e-156
Identities = 264/386 (68%), Positives = 320/386 (82%), Gaps = 14/386 (3%)
Query: 11 ESSDFTSEDEGTEDYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYV 70
+ S+++SEDEGTEDY++GGYH VR+GDTF +G YV+QSKLGWGHFSTVWLAWD+ SRYV
Sbjct: 12 DESEYSSEDEGTEDYKKGGYHTVRVGDTFKNGAYVIQSKLGWGHFSTVWLAWDTQESRYV 71
Query: 71 ALKVQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYL 130
ALKVQKSAQHYTEAA+DEI IL+QIAEGD+ DKKCVVKLLDHFKH+GPNG+HVCMVFEYL
Sbjct: 72 ALKVQKSAQHYTEAAMDEIKILKQIAEGDSGDKKCVVKLLDHFKHTGPNGKHVCMVFEYL 131
Query: 131 GDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLSTIDPS 190
GDNLL++IKYSDYRG+P++MVKE+CFHILVGLDYLH++LSIIHTDLKPEN+LLLSTIDPS
Sbjct: 132 GDNLLSVIKYSDYRGVPLHMVKELCFHILVGLDYLHRELSIIHTDLKPENVLLLSTIDPS 191
Query: 191 KDPRKSGAPLILPNSKDKTMLESTAARDTK--TSNGDFIKNHKKNIKRKAKQ-AAHGCAE 247
+D R+SG PL+LP +KDK + ES +TK T NGD KN KK I++KAK+ A
Sbjct: 192 RDVRRSGVPLVLPITKDKIVSESAVKPETKSYTYNGDLTKNQKKKIRKKAKKVVAQDFGG 251
Query: 248 KEASEGVDGNHETSGAVESSPNASSAREQASSSAGTSRLSDADGMKLKE-----QGNRRG 302
+EA E + +++ + N++ R + SS +RL + + + K +G+RRG
Sbjct: 252 EEALE--ESERDSNSEARINGNSTVERSEGSS----TRLMEGEEAREKANKKNGRGSRRG 305
Query: 303 SRTVRQKLLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSF 362
SR+ RQKLL+ + KCKLVDFGNACWTYKQFT+DIQTRQYRCPEV+LGSKYSTSAD+WSF
Sbjct: 306 SRSTRQKLLSDIECKCKLVDFGNACWTYKQFTSDIQTRQYRCPEVVLGSKYSTSADMWSF 365
Query: 363 ACICFELATGDVLFDPHSGDNFDRDE 388
ACICFELATGDVLFDPHSG+N+DRDE
Sbjct: 366 ACICFELATGDVLFDPHSGENYDRDE 391
>At4g35500 protein kinase - like protein
Length = 438
Score = 356 bits (914), Expect = 1e-98
Identities = 191/379 (50%), Positives = 236/379 (61%), Gaps = 56/379 (14%)
Query: 10 TESSDFTSEDEGTEDYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRY 69
+ SS E+EG + YR+GGYHAVRIGD FS GRY+ Q KLGWG FSTVWLA+D+ S Y
Sbjct: 4 SSSSGSEGEEEGFDAYRKGGYHAVRIGDPFSGGRYIAQRKLGWGQFSTVWLAYDTLTSTY 63
Query: 70 VALKVQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEY 129
VALK+QKSAQ + +AAL EI L A+GD D KCVV+L+DHFKHSGPNGQH+CMV E+
Sbjct: 64 VALKIQKSAQQFAQAALHEIEFLSAAADGDLDKTKCVVRLIDHFKHSGPNGQHLCMVLEF 123
Query: 130 LGDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLSTIDP 189
LGD+LL LI+Y+ Y+G+ +N V+EIC IL GLDYLH++L +IH+DLKPENILL STIDP
Sbjct: 124 LGDSLLRLIRYNQYKGLKLNKVREICRCILTGLDYLHRELGMIHSDLKPENILLCSTIDP 183
Query: 190 SKDPRKSGAPLILPNSKDKTMLESTAARDTKTSNGDFIKNHKKNIKRKAKQAAHGCAEKE 249
+KDP +SG +L E A T N +K +KR+AK+A +E+
Sbjct: 184 AKDPVRSGLTPLLEKP------EGNANGGASTMN-----LIEKKLKRRAKRAVAKISERR 232
Query: 250 ASEGVDGNHETSGAVESSPNASSAREQASSSAGTSRLSDADGMKLKEQGNRRGSRTVRQK 309
S V G E+ASS +K
Sbjct: 233 VSM-VTG------------------EEASSKT--------------------------EK 247
Query: 310 LLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFEL 369
L D++CK+VDFGNACW KQF +IQTRQYR PEVIL S YS S D+WSF C FEL
Sbjct: 248 SLDGIDMRCKVVDFGNACWADKQFAEEIQTRQYRAPEVILKSGYSFSVDMWSFGCTAFEL 307
Query: 370 ATGDVLFDPHSGDNFDRDE 388
TGD+LF P G+ + DE
Sbjct: 308 VTGDMLFAPKDGNGYGEDE 326
>At2g17530 putative protein kinase
Length = 440
Score = 347 bits (891), Expect = 5e-96
Identities = 185/379 (48%), Positives = 237/379 (61%), Gaps = 57/379 (15%)
Query: 10 TESSDFTSEDEGTEDYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRY 69
+ SS +DEG + YR+GGYHAVRIGD F+ GRY+ Q KLGWG FSTVWLA+D+ S Y
Sbjct: 4 SSSSGSEEDDEGFDAYRKGGYHAVRIGDQFAGGRYIAQRKLGWGQFSTVWLAYDTRTSNY 63
Query: 70 VALKVQKSAQHYTEAALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEY 129
VALK+QKSA + +AAL EI +LQ A+GD ++ KCV++L+D FKH+GPNGQH+CMV E+
Sbjct: 64 VALKIQKSALQFAQAALHEIELLQAAADGDPENTKCVIRLIDDFKHAGPNGQHLCMVLEF 123
Query: 130 LGDNLLTLIKYSDYRGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLSTIDP 189
LGD+LL LIKY+ Y+GM ++ V+EIC IL GLDYLH++L +IH+DLKPENILL STIDP
Sbjct: 124 LGDSLLRLIKYNRYKGMELSKVREICKCILTGLDYLHRELGMIHSDLKPENILLCSTIDP 183
Query: 190 SKDPRKSGAPLILPNSKDKTMLESTAARDTKTSNGDFIKNHKKNIKRKAKQAAHGCAEKE 249
+KDP +SG I LE TS + I +K +KR+AK+AA + +
Sbjct: 184 AKDPIRSGLTPI---------LEKPEGNQNGTSTMNLI---EKKLKRRAKKAAAKISGRR 231
Query: 250 ASEGVDGNHETSGAVESSPNASSAREQASSSAGTSRLSDADGMKLKEQGNRRGSRTVRQK 309
S + G ET + N+R
Sbjct: 232 VS--IVGLSET-----------------------------------PKKNKRN------- 247
Query: 310 LLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFEL 369
L D++CK+VDFGN CW +F +IQTRQYR PEVIL S YS S D+WSFAC FEL
Sbjct: 248 -LDGIDMRCKVVDFGNGCWADNKFAEEIQTRQYRAPEVILQSGYSYSVDMWSFACTAFEL 306
Query: 370 ATGDVLFDPHSGDNFDRDE 388
ATGD+LF P G+ + DE
Sbjct: 307 ATGDMLFAPKEGNGYGEDE 325
>At4g24740 protein kinase (AFC2)
Length = 427
Score = 113 bits (283), Expect = 2e-25
Identities = 65/174 (37%), Positives = 104/174 (59%), Gaps = 8/174 (4%)
Query: 27 RGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAAL 86
+ G++ +GD + RY + SK+G G F V WD VA+K+ + + Y EAA+
Sbjct: 82 KDGHYIFELGDDLTP-RYKIYSKMGEGTFGQVLECWDRERKEMVAVKIVRGVKKYREAAM 140
Query: 87 DEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGM 146
EI +LQQ+ + D +C V++ + F + H+C+VFE LG +L ++ ++YR
Sbjct: 141 IEIEMLQQLGKHDKGGNRC-VQIRNWFDYR----NHICIVFEKLGSSLYDFLRKNNYRSF 195
Query: 147 PINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLSTIDPSKDPRKSGAPL 200
PI++V+EI + +L + ++H L +IHTDLKPENILL+S+ D K P G+ L
Sbjct: 196 PIDLVREIGWQLLECVAFMH-DLRMIHTDLKPENILLVSS-DYVKIPEYKGSRL 247
Score = 65.9 bits (159), Expect = 4e-11
Identities = 36/94 (38%), Positives = 50/94 (52%), Gaps = 1/94 (1%)
Query: 286 LSDADGMKLKEQGNRRGSRTVRQKLLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCP 345
L +D +K+ E R R V K + + K++DFG+ + + T + TR YR P
Sbjct: 231 LVSSDYVKIPEYKGSRLQRDVCYKRVPKSSA-IKVIDFGSTTYERQDQTYIVSTRHYRAP 289
Query: 346 EVILGSKYSTSADLWSFACICFELATGDVLFDPH 379
EVILG +S D+WS CI EL TG+ LF H
Sbjct: 290 EVILGLGWSYPCDVWSVGCIIVELCTGEALFQTH 323
>At3g53570 protein kinase (AME2/AFC1)
Length = 467
Score = 110 bits (275), Expect = 1e-24
Identities = 61/160 (38%), Positives = 98/160 (61%), Gaps = 7/160 (4%)
Query: 27 RGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAAL 86
+ G++ +GDT + RY + SK+G G F V +D+ + VA+KV +S Y EAA+
Sbjct: 99 KDGHYVFVVGDTLTP-RYQILSKMGEGTFGQVLECFDNKNKEVVAIKVIRSINKYREAAM 157
Query: 87 DEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGM 146
EI +LQ++ D +C V++ + F + H+C+VFE LG +L ++ + YR
Sbjct: 158 IEIDVLQRLTRHDVGGSRC-VQIRNWFDYR----NHICIVFEKLGPSLYDFLRKNSYRSF 212
Query: 147 PINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLST 186
PI++V+E+ +L + Y+H L +IHTDLKPENILL+S+
Sbjct: 213 PIDLVRELGRQLLESVAYMH-DLRLIHTDLKPENILLVSS 251
Score = 62.0 bits (149), Expect = 5e-10
Identities = 27/61 (44%), Positives = 37/61 (60%)
Query: 319 KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFDP 378
KL+DFG+ + ++ + TR YR PEVILG ++ DLWS CI EL +G+ LF
Sbjct: 282 KLIDFGSTTFEHQDHNYIVSTRHYRAPEVILGVGWNYPCDLWSIGCILVELCSGEALFQT 341
Query: 379 H 379
H
Sbjct: 342 H 342
>At4g32660 protein kinase (AME3)
Length = 400
Score = 107 bits (267), Expect = 1e-23
Identities = 58/163 (35%), Positives = 93/163 (56%), Gaps = 7/163 (4%)
Query: 24 DYRRGGYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTE 83
D R G++ + D + RY + SK+G G F V WD YVA+K+ +S + Y +
Sbjct: 52 DDDRDGHYVFSLRDNLTP-RYKILSKMGEGTFGRVLECWDRDTKEYVAIKIIRSIKKYRD 110
Query: 84 AALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDY 143
AA+ EI +LQ++ + D +CV K+ H+C+VFE LG +L +K + Y
Sbjct: 111 AAMIEIDVLQKLVKSDKGRTRCV-----QMKNWFDYRNHICIVFEKLGPSLFDFLKRNKY 165
Query: 144 RGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLST 186
P+ +V++ +L + Y+H +L ++HTDLKPENILL+S+
Sbjct: 166 SAFPLALVRDFGCQLLESVAYMH-ELQLVHTDLKPENILLVSS 207
Score = 67.8 bits (164), Expect = 1e-11
Identities = 39/100 (39%), Positives = 55/100 (55%), Gaps = 4/100 (4%)
Query: 286 LSDADGMKLKEQGNRRGSRTVRQKLLASADVKCKLVDFGNACWTYKQFTNDIQTRQYRCP 345
L ++ +KL + + T + L S+ +K L+DFG+ + + +QTR YR P
Sbjct: 204 LVSSENVKLPDNKRSAANETHFRCLPKSSAIK--LIDFGSTVCDNRIHHSIVQTRHYRSP 261
Query: 346 EVILGSKYSTSADLWSFACICFELATGDVLFDPHSGDNFD 385
EVILG +S DLWS CI FEL TG+ LF H DN +
Sbjct: 262 EVILGLGWSYQCDLWSIGCILFELCTGEALFQTH--DNLE 299
>At5g35980 protein kinase-like
Length = 787
Score = 103 bits (257), Expect = 2e-22
Identities = 63/162 (38%), Positives = 94/162 (57%), Gaps = 10/162 (6%)
Query: 35 IGDTFSSG----RYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEIT 90
+ D F S RY+V+ LG G F V W + +VA+KV K+ Y + AL E++
Sbjct: 109 VNDDFCSSDSRQRYIVKDLLGHGTFGQVAKCWVPETNSFVAVKVIKNQLAYYQQALVEVS 168
Query: 91 ILQQIAEG-DTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGMPIN 149
IL + + D +DK +V++ D+F H H+C+ FE L NL LIK + +RG+ ++
Sbjct: 169 ILTTLNKKYDPEDKNHIVRIYDYFLHQS----HLCICFELLDMNLYELIKINQFRGLSLS 224
Query: 150 MVKEICFHILVGLDYLHKQLSIIHTDLKPENILLLSTIDPSK 191
+VK IL+GL L K IIH DLKPENILL +++ P++
Sbjct: 225 IVKLFSKQILLGLALL-KDAGIIHCDLKPENILLCASVKPTE 265
Score = 67.8 bits (164), Expect = 1e-11
Identities = 37/81 (45%), Positives = 49/81 (59%), Gaps = 5/81 (6%)
Query: 308 QKLLASADVK---CKLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFAC 364
+ +L A VK K++DFG+AC K + IQ+R YR PEV+LG +Y+T+ D+WSF C
Sbjct: 253 ENILLCASVKPTEIKIIDFGSACMEDKTVYSYIQSRYYRSPEVLLGYQYTTAIDMWSFGC 312
Query: 365 ICFELATGDVLFDPHSGDNFD 385
I EL G LF G FD
Sbjct: 313 IVAELFLGLPLFP--GGSEFD 331
>At2g40120 protein kinase like protein
Length = 570
Score = 77.4 bits (189), Expect = 1e-14
Identities = 52/153 (33%), Positives = 82/153 (52%), Gaps = 9/153 (5%)
Query: 37 DTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIA 96
+T GRY + +G FS V A D H+ V LK+ K+ + + + +LDEI +L+ +
Sbjct: 254 NTVIGGRYYITEYIGSAAFSKVVQAQDLHNGVDVCLKIIKNDKDFFDQSLDEIKLLKHVN 313
Query: 97 EGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRG----MPINMVK 152
+ D D+ +++L D+F H +H+ +V E L NL K++ G ++ ++
Sbjct: 314 KHDPADEHHILRLYDYFYHQ----EHLFIVCELLRANLYEFQKFNQESGGEPYFNLSRLQ 369
Query: 153 EICFHILVGLDYLHKQLSIIHTDLKPENILLLS 185
I L L +LH L IIH DLKPENIL+ S
Sbjct: 370 VITRQCLDALVFLH-GLGIIHCDLKPENILIKS 401
Score = 62.0 bits (149), Expect = 5e-10
Identities = 29/71 (40%), Positives = 42/71 (58%), Gaps = 2/71 (2%)
Query: 308 QKLLASADVKC--KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACI 365
+ +L + +C K++D G++C+ +Q+R YR PEVILG Y DLWS CI
Sbjct: 395 ENILIKSYKRCAVKIIDLGSSCFRSDNLCLYVQSRSYRAPEVILGLPYDEKIDLWSLGCI 454
Query: 366 CFELATGDVLF 376
EL +G+VLF
Sbjct: 455 LAELCSGEVLF 465
>At1g13350 Unknown protein
Length = 761
Score = 73.9 bits (180), Expect = 1e-13
Identities = 49/160 (30%), Positives = 88/160 (54%), Gaps = 11/160 (6%)
Query: 29 GYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSH----HSRYVALKVQKSAQHYTEA 84
GY++ ++G+ RY + + G G FSTV A D+ VA+K+ ++ + +A
Sbjct: 428 GYYSYQLGELLDD-RYEIMATHGKGVFSTVVRAKDTKAELGEPEEVAIKIIRNNETMHKA 486
Query: 85 ALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIK-YSDY 143
EI IL+++A D ++K+ V+ L FK+ H+C+VFE L NL ++K Y
Sbjct: 487 GQTEIQILKKLAGSDPENKRHCVRFLSTFKYRN----HLCLVFESLHLNLREIVKKYGRN 542
Query: 144 RGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILL 183
G+ ++ V+ + + L +L K ++H D+KP+N+L+
Sbjct: 543 IGIQLSGVRVYATQLFISLKHL-KNCGVLHCDIKPDNMLV 581
Score = 50.8 bits (120), Expect = 1e-06
Identities = 27/66 (40%), Positives = 39/66 (58%), Gaps = 2/66 (3%)
Query: 319 KLVDFGNACWT-YKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFD 377
KL DFG+A + + T + +R YR PE+ILG Y D+WS C +EL +G ++F
Sbjct: 589 KLCDFGSAMFAGTNEVTPYLVSRFYRAPEIILGLPYDHPLDIWSVGCCLYELFSGKIMF- 647
Query: 378 PHSGDN 383
P S +N
Sbjct: 648 PGSTNN 653
>At3g25840 protein kinase like protein
Length = 935
Score = 72.0 bits (175), Expect = 5e-13
Identities = 49/160 (30%), Positives = 87/160 (53%), Gaps = 11/160 (6%)
Query: 29 GYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSH----HSRYVALKVQKSAQHYTEA 84
GY++ + G+ GRY V + G G FSTV A D VA+K+ ++ + +A
Sbjct: 603 GYYSYQFGELLD-GRYEVIATHGKGVFSTVVRAKDLKAGPAEPEEVAIKIIRNNETMHKA 661
Query: 85 ALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIK-YSDY 143
E+ IL+++A D +D++ V+ L FK+ H+C+VFE L NL ++K +
Sbjct: 662 GKIEVQILKKLAGADREDRRHCVRFLSSFKYRN----HLCLVFESLHLNLREVLKKFGRN 717
Query: 144 RGMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILL 183
G+ ++ V+ + + L +L K ++H D+KP+N+L+
Sbjct: 718 IGLQLSAVRAYSKQLFIALKHL-KNCGVLHCDIKPDNMLV 756
Score = 54.3 bits (129), Expect = 1e-07
Identities = 28/59 (47%), Positives = 36/59 (60%), Gaps = 1/59 (1%)
Query: 319 KLVDFGNACWTYK-QFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLF 376
KL DFGNA + K + T + +R YR PE+ILG Y D+WS C +EL +G VLF
Sbjct: 764 KLCDFGNAMFAGKNEVTPYLVSRFYRSPEIILGLTYDHPLDIWSVGCCLYELYSGKVLF 822
>At3g17750 protein kinase, putative
Length = 1138
Score = 71.6 bits (174), Expect = 7e-13
Identities = 50/149 (33%), Positives = 78/149 (51%), Gaps = 9/149 (6%)
Query: 41 SGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDT 100
+GRY V LG FS A D H V +K+ K+ + + + +LDEI +L+ + + D
Sbjct: 824 AGRYHVTEHLGSAAFSKAIQAHDLHTGIDVCVKIIKNNKDFFDQSLDEIKLLKYVNQHDP 883
Query: 101 DDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRG----MPINMVKEICF 156
DK +++L D+F +H+ +V E L NL K++ G + ++ I
Sbjct: 884 ADKYHLLRLYDYFYFR----EHLLIVCELLKANLYEFQKFNRESGGEVYFTMPRLQSITI 939
Query: 157 HILVGLDYLHKQLSIIHTDLKPENILLLS 185
L L++LH L +IH DLKPENIL+ S
Sbjct: 940 QCLEALNFLH-GLGLIHCDLKPENILIKS 967
Score = 63.9 bits (154), Expect = 1e-10
Identities = 30/75 (40%), Positives = 44/75 (58%), Gaps = 2/75 (2%)
Query: 308 QKLLASADVKC--KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACI 365
+ +L + +C K++D G++C+ + +Q+R YR PEVILG Y D+WS CI
Sbjct: 961 ENILIKSYSRCEIKVIDLGSSCFETDHLCSYVQSRSYRAPEVILGLPYDKKIDIWSLGCI 1020
Query: 366 CFELATGDVLFDPHS 380
EL TG+VLF S
Sbjct: 1021 LAELCTGNVLFQNDS 1035
>At1g73450
Length = 1155
Score = 69.3 bits (168), Expect = 3e-12
Identities = 49/149 (32%), Positives = 77/149 (50%), Gaps = 9/149 (6%)
Query: 41 SGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDT 100
+GRY V LG FS A D V +K+ K+ + + + +LDEI +L+ + + D
Sbjct: 826 AGRYHVTEYLGSAAFSKAIQAHDLQTGMDVCIKIIKNNKDFFDQSLDEIKLLKYVNKHDP 885
Query: 101 DDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRG----MPINMVKEICF 156
DK +++L D+F + +H+ +V E L NL K++ G + ++ I
Sbjct: 886 ADKYHLLRLYDYFYYR----EHLLIVCELLKANLYEFHKFNRESGGEVYFTMPRLQSITI 941
Query: 157 HILVGLDYLHKQLSIIHTDLKPENILLLS 185
L L +LH L +IH DLKPENIL+ S
Sbjct: 942 QCLESLQFLH-GLGLIHCDLKPENILVKS 969
Score = 58.2 bits (139), Expect = 8e-09
Identities = 26/68 (38%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query: 308 QKLLASADVKC--KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACI 365
+ +L + +C K++D G++C+ + +Q+R YR PEVILG Y D+WS CI
Sbjct: 963 ENILVKSYSRCEIKVIDLGSSCFETDHLCSYVQSRSYRAPEVILGLPYDKKIDVWSLGCI 1022
Query: 366 CFELATGD 373
EL TG+
Sbjct: 1023 LAELCTGN 1030
>At1g73460
Length = 1157
Score = 69.3 bits (168), Expect = 3e-12
Identities = 49/149 (32%), Positives = 77/149 (50%), Gaps = 9/149 (6%)
Query: 41 SGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDT 100
+GRY V LG FS A D V +K+ K+ + + + +LDEI +L+ + + D
Sbjct: 843 AGRYHVTEYLGSAAFSKAIQAHDLQTGMDVCIKIIKNNKDFFDQSLDEIKLLKYVNKHDP 902
Query: 101 DDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRG----MPINMVKEICF 156
DK +++L D+F + +H+ +V E L NL K++ G + ++ I
Sbjct: 903 ADKYHLLRLYDYFYYR----EHLLIVCELLKANLYEFHKFNRESGGEVYFTMPRLQSITI 958
Query: 157 HILVGLDYLHKQLSIIHTDLKPENILLLS 185
L L +LH L +IH DLKPENIL+ S
Sbjct: 959 QCLESLQFLH-GLGLIHCDLKPENILVKS 986
Score = 63.9 bits (154), Expect = 1e-10
Identities = 30/75 (40%), Positives = 44/75 (58%), Gaps = 2/75 (2%)
Query: 308 QKLLASADVKC--KLVDFGNACWTYKQFTNDIQTRQYRCPEVILGSKYSTSADLWSFACI 365
+ +L + +C K++D G++C+ + +Q+R YR PEVILG Y D+WS CI
Sbjct: 980 ENILVKSYSRCEIKVIDLGSSCFETDHLCSYVQSRSYRAPEVILGLPYDKKIDVWSLGCI 1039
Query: 366 CFELATGDVLFDPHS 380
EL TG+VLF S
Sbjct: 1040 LAELCTGNVLFQNDS 1054
>At3g53640 protein kinase -like protein
Length = 642
Score = 67.4 bits (163), Expect = 1e-11
Identities = 48/160 (30%), Positives = 84/160 (52%), Gaps = 11/160 (6%)
Query: 29 GYHAVRIGDTFSSGRYVVQSKLGWGHFSTVWLAWDSH----HSRYVALKVQKSAQHYTEA 84
GY++ ++G+ RY + + G G FSTV A D+ VA+K+ + + +A
Sbjct: 309 GYYSYQLGELLDD-RYEIMATHGKGVFSTVVRAKDTKPELGEPEEVAIKIIRKNETMHKA 367
Query: 85 ALDEITILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYR 144
EI IL+++ D ++K V+LL F++ H+C+VFE L NL ++K
Sbjct: 368 GQAEIRILKKLVCSDPENKHHCVRLLSTFEYRN----HLCLVFESLHLNLREVVKKIGVN 423
Query: 145 -GMPINMVKEICFHILVGLDYLHKQLSIIHTDLKPENILL 183
G+ + V+ + + L +L K ++H D+KP+NIL+
Sbjct: 424 IGLKLYDVRVYAEQLFISLKHL-KNCGVLHCDIKPDNILM 462
Score = 52.0 bits (123), Expect = 6e-07
Identities = 28/66 (42%), Positives = 40/66 (60%), Gaps = 2/66 (3%)
Query: 319 KLVDFGNACWTYK-QFTNDIQTRQYRCPEVILGSKYSTSADLWSFACICFELATGDVLFD 377
KL DFG+A + + Q T + +R YR PE+ILG Y D+WS C +EL +G ++F
Sbjct: 470 KLCDFGSAMFAGENQVTPYLVSRFYRAPEIILGLPYDHPLDIWSVGCCLYELYSGKIMF- 528
Query: 378 PHSGDN 383
P S +N
Sbjct: 529 PGSTNN 534
>At1g66750 unknown protein
Length = 348
Score = 57.4 bits (137), Expect = 1e-08
Identities = 47/153 (30%), Positives = 78/153 (50%), Gaps = 19/153 (12%)
Query: 36 GDTFSSGRYVVQSKLGWGHFSTVWLAWDSHHSRYVALKV-----QKSAQHYTEAALDEIT 90
GD RY+ + LG G + V+ A D+ + VA+K QK ++T AL EI
Sbjct: 5 GDNQPVDRYLRRQILGEGTYGVVYKATDTKTGKTVAVKKIRLGNQKEGVNFT--ALREIK 62
Query: 91 ILQQIAEGDTDDKKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGMPINM 150
+L+++ + +V+L+D F H G + +VFEY+ +L +I+ + P
Sbjct: 63 LLKEL------NHPHIVELIDAFPHDG----SLHLVFEYMQTDLEAVIRDRNIFLSP-GD 111
Query: 151 VKEICFHILVGLDYLHKQLSIIHTDLKPENILL 183
+K L GL Y HK+ ++H D+KP N+L+
Sbjct: 112 IKSYMLMTLKGLAYCHKKW-VLHRDMKPNNLLI 143
Score = 38.5 bits (88), Expect = 0.006
Identities = 23/64 (35%), Positives = 33/64 (50%), Gaps = 4/64 (6%)
Query: 310 LLASADVKCKLVDFGNACW---TYKQFTNDIQTRQYRCPEVILGSK-YSTSADLWSFACI 365
LL + KL DFG A ++FT+ + YR PE++ GS+ Y D+W+ CI
Sbjct: 141 LLIGENGLLKLADFGLARLFGSPNRRFTHQVFATWYRAPELLFGSRQYGAGVDVWAAGCI 200
Query: 366 CFEL 369
EL
Sbjct: 201 FAEL 204
>At4g19110 putative serine/threonine-protein kinase
Length = 461
Score = 55.8 bits (133), Expect = 4e-08
Identities = 43/145 (29%), Positives = 68/145 (46%), Gaps = 10/145 (6%)
Query: 43 RYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDTDD 102
RY + ++G G F +VW A + VA+K K + + DE L+++ +
Sbjct: 3 RYKLIKEVGDGTFGSVWRAINKQTGEVVAIKKMKKKYY----SWDECINLREVKSLRRMN 58
Query: 103 KKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGMPINMVKEICFHILVGL 162
+VKL + + + + VFEY+ NL L+K + +K CF + GL
Sbjct: 59 HPNIVKLKEVIREN----DILYFVFEYMECNLYQLMKDRQKLFAEAD-IKNWCFQVFQGL 113
Query: 163 DYLHKQLSIIHTDLKPENILLLSTI 187
Y+H Q H DLKPEN+L+ I
Sbjct: 114 SYMH-QRGYFHRDLKPENLLVSKDI 137
Score = 40.8 bits (94), Expect = 0.001
Identities = 26/65 (40%), Positives = 35/65 (53%), Gaps = 4/65 (6%)
Query: 308 QKLLASADVKCKLVDFGNACWTYKQ--FTNDIQTRQYRCPEVILGS-KYSTSADLWSFAC 364
+ LL S D+ K+ DFG A FT + TR YR PEV+L S Y++ D+W+
Sbjct: 129 ENLLVSKDI-IKIADFGLAREVNSSPPFTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGA 187
Query: 365 ICFEL 369
I EL
Sbjct: 188 IMAEL 192
>At3g07980 putative MAP3K epsilon protein kinase
Length = 1367
Score = 55.1 bits (131), Expect = 7e-08
Identities = 42/141 (29%), Positives = 76/141 (53%), Gaps = 9/141 (6%)
Query: 43 RYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDTDD 102
+Y++ ++G G + V++ D + +VA+K Q S ++ + L+ TI+Q+I +
Sbjct: 19 KYMLGDEIGKGAYGRVYIGLDLENGDFVAIK-QVSLENIGQEDLN--TIMQEIDLLKNLN 75
Query: 103 KKCVVKLLDHFKHSGPNGQHVCMVFEYLGD-NLLTLIKYSDYRGMPINMVKEICFHILVG 161
K +VK L K H+ ++ EY+ + +L +IK + + P ++V +L G
Sbjct: 76 HKNIVKYLGSLKTK----THLHIILEYVENGSLANIIKPNKFGPFPESLVTVYIAQVLEG 131
Query: 162 LDYLHKQLSIIHTDLKPENIL 182
L YLH+Q +IH D+K NIL
Sbjct: 132 LVYLHEQ-GVIHRDIKGANIL 151
Score = 31.6 bits (70), Expect = 0.79
Identities = 21/65 (32%), Positives = 30/65 (45%), Gaps = 3/65 (4%)
Query: 310 LLASADVKCKLVDFGNACWTYKQFTND---IQTRQYRCPEVILGSKYSTSADLWSFACIC 366
+L + + KL DFG A + N + T + PEVI S ++D+WS C
Sbjct: 150 ILTTKEGLVKLADFGVATKLNEADFNTHSVVGTPYWMAPEVIELSGVCAASDIWSVGCTI 209
Query: 367 FELAT 371
EL T
Sbjct: 210 IELLT 214
>At5g45430 serine/threonine-protein kinase Mak (male germ
cell-associated kinase)-like protein
Length = 499
Score = 54.3 bits (129), Expect = 1e-07
Identities = 40/141 (28%), Positives = 67/141 (47%), Gaps = 10/141 (7%)
Query: 43 RYVVQSKLGWGHFSTVWLAWDSHHSRYVALKVQKSAQHYTEAALDEITILQQIAEGDTDD 102
RY + ++G G F VW A + + VA+K K + +E L+++ +
Sbjct: 3 RYTLLKEVGDGTFGNVWRAVNKQTNEVVAIKRMKKKYF----SWEECVNLREVKSLSRMN 58
Query: 103 KKCVVKLLDHFKHSGPNGQHVCMVFEYLGDNLLTLIKYSDYRGMPINMVKEICFHILVGL 162
+VKL + + + + VFEY+ NL L+K + + ++ CF + GL
Sbjct: 59 HPNIVKLKEVIREN----DILYFVFEYMECNLYQLMK-DRPKHFAESDIRNWCFQVFQGL 113
Query: 163 DYLHKQLSIIHTDLKPENILL 183
Y+H Q H DLKPEN+L+
Sbjct: 114 SYMH-QRGYFHRDLKPENLLV 133
Score = 38.5 bits (88), Expect = 0.006
Identities = 27/72 (37%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query: 308 QKLLASADVKCKLVDFGNA--CWTYKQFTNDIQTRQYRCPEVILGS-KYSTSADLWSFAC 364
+ LL S DV K+ D G A + +T + TR YR PEV+L S Y++ D+W+
Sbjct: 129 ENLLVSKDV-IKIADLGLAREIDSSPPYTEYVSTRWYRAPEVLLQSYVYTSKVDMWAMGA 187
Query: 365 ICFELATGDVLF 376
I EL + LF
Sbjct: 188 IMAELLSLRPLF 199
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,300,171
Number of Sequences: 26719
Number of extensions: 399871
Number of successful extensions: 3013
Number of sequences better than 10.0: 788
Number of HSP's better than 10.0 without gapping: 283
Number of HSP's successfully gapped in prelim test: 506
Number of HSP's that attempted gapping in prelim test: 2075
Number of HSP's gapped (non-prelim): 1328
length of query: 395
length of database: 11,318,596
effective HSP length: 101
effective length of query: 294
effective length of database: 8,619,977
effective search space: 2534273238
effective search space used: 2534273238
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC139525.1


