

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139355.5 - phase: 0
(672 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
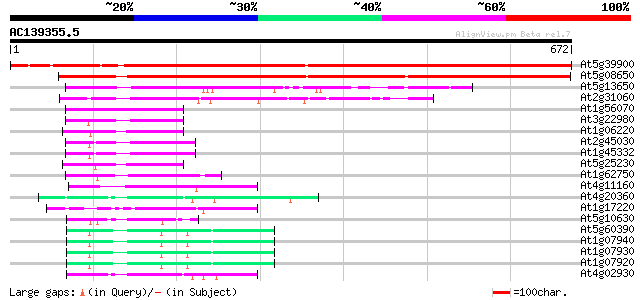
Score E
Sequences producing significant alignments: (bits) Value
At5g39900 GTP-binding protein-like 1024 0.0
At5g08650 GTP-binding protein LepA homolog 612 e-175
At5g13650 GTP-binding protein typA (tyrosine phosphorylated prot... 148 1e-35
At2g31060 putative GTP-binding protein 129 5e-30
At1g56070 elongation factor like protein 105 6e-23
At3g22980 eukaryotic translation elongation factor 2, putative 96 6e-20
At1g06220 elongation factor like protein 95 1e-19
At2g45030 putative mitochondrial translation elongation factor G 92 7e-19
At1g45332 mitochondrial elongation factor, putative 92 7e-19
At5g25230 translation Elongation Factor 2 - like protein 91 3e-18
At1g62750 elongation factor G, putative 82 1e-15
At4g11160 translation initiation factor IF-2 like protein 74 2e-13
At4g20360 translation elongation factor EF-Tu precursor, chlorop... 67 2e-11
At1g17220 putative translation initiation factor IF2 64 3e-10
At5g10630 putative protein 59 9e-09
At5g60390 translation elongation factor eEF-1 alpha chain (gene A4) 59 1e-08
At1g07940 elongation factor 1-alpha 59 1e-08
At1g07930 elongation factor 1-alpha 59 1e-08
At1g07920 elongation factor 1-alpha 59 1e-08
At4g02930 mitochondrial elongation factor Tu 58 2e-08
>At5g39900 GTP-binding protein-like
Length = 661
Score = 1024 bits (2648), Expect = 0.0
Identities = 520/674 (77%), Positives = 595/674 (88%), Gaps = 15/674 (2%)
Query: 1 MGFLRKASKTLKQSNY-VSLLFNFNPLSSRITHERFSITRAL-FCTQSRQNYTKEKAIID 58
MG + +ASKTLK S +S+LFN + S+R + +A F + SRQ+ ++ ID
Sbjct: 1 MGSMYRASKTLKSSRQALSILFN-SLNSNRQNPTCIGLYQAYGFSSDSRQS--SKEPTID 57
Query: 59 LSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVK 118
L+++P E +RNFSIIAH+DHGKSTLADRL+ELTGTIKKG GQPQYLDKLQ RERGITVK
Sbjct: 58 LTKFPSEKIRNFSIIAHIDHGKSTLADRLMELTGTIKKGHGQPQYLDKLQ--RERGITVK 115
Query: 119 AQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAA 178
AQTATMFY+N + + +E+S YLLNLIDTPGHVDFSYEVSRSL+ACQG LLVVDAA
Sbjct: 116 AQTATMFYENKV------EDQEASGYLLNLIDTPGHVDFSYEVSRSLSACQGALLVVDAA 169
Query: 179 QGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKT 238
QGVQAQTVANFYLAFE+NL I+PVINKIDQPTADP+RVK QLKSMFDLD D LL SAKT
Sbjct: 170 QGVQAQTVANFYLAFEANLTIVPVINKIDQPTADPERVKAQLKSMFDLDTEDVLLVSAKT 229
Query: 239 GVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISS 298
G+GLEHVLPAVIERIPPPPG SES LRMLL DS+F+EY+GVIC+V+VVDG L KGDK+S
Sbjct: 230 GLGLEHVLPAVIERIPPPPGISESPLRMLLFDSFFNEYKGVICYVSVVDGMLSKGDKVSF 289
Query: 299 AATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEP 358
AA+G+SYE +D+GIMHPELT TG+L TGQVGY++TGMRTTKEARIGDTIY TK+TV EP
Sbjct: 290 AASGQSYEVLDVGIMHPELTSTGMLLTGQVGYIVTGMRTTKEARIGDTIYRTKTTV--EP 347
Query: 359 LPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLG 418
LPGFK +HMVFSG++PADGSDFEAL HA+EKLTCNDASVSV KETSTALG+GFRCGFLG
Sbjct: 348 LPGFKPVRHMVFSGVYPADGSDFEALGHAMEKLTCNDASVSVAKETSTALGMGFRCGFLG 407
Query: 419 LLHMDVFHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEP 478
LLHMDVFHQRLEQEYG +IST+PTVPY +EYSDGSKL+VQNPAALPSNPK RV A WEP
Sbjct: 408 LLHMDVFHQRLEQEYGTQVISTIPTVPYTFEYSDGSKLQVQNPAALPSNPKYRVTASWEP 467
Query: 479 TVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSI 538
TVIATI++PSEYVG VI L S+RRG+QLEY+FID+QRVF+KY+LPLREIV+DFY+ELKSI
Sbjct: 468 TVIATIILPSEYVGAVINLCSDRRGQQLEYTFIDAQRVFLKYQLPLREIVVDFYDELKSI 527
Query: 539 TSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDR 598
TSGYASFDYED++YQ SDLVKLDILLNGQAVDA+ATIVH K+YRVG+ELVEKLK I+R
Sbjct: 528 TSGYASFDYEDAEYQASDLVKLDILLNGQAVDALATIVHKQKAYRVGKELVEKLKNYIER 587
Query: 599 QMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 658
QMFE++IQAAIGSKIIAR+T++AMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV
Sbjct: 588 QMFEVMIQAAIGSKIIARDTISAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 647
Query: 659 DVPQEAFHELLKVS 672
D+P EAF ++LKVS
Sbjct: 648 DIPHEAFQQILKVS 661
>At5g08650 GTP-binding protein LepA homolog
Length = 675
Score = 612 bits (1578), Expect = e-175
Identities = 313/613 (51%), Positives = 425/613 (69%), Gaps = 14/613 (2%)
Query: 59 LSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVK 118
L + P +RNFSIIAH+DHGKSTLAD+LL++TGT++ + Q+LD + +ERERGIT+K
Sbjct: 72 LLKVPISNIRNFSIIAHIDHGKSTLADKLLQVTGTVQNRDMKEQFLDNMDLERERGITIK 131
Query: 119 AQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAA 178
Q A M Y E + + LNLIDTPGHVDFSYEVSRSLAAC+G LLVVDA+
Sbjct: 132 LQAARMRYVY-----------EDTPFCLNLIDTPGHVDFSYEVSRSLAACEGALLVVDAS 180
Query: 179 QGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKT 238
QGV+AQT+AN YLA E+NL IIPV+NKID P A+P++V +++ + LD S A+ SAK
Sbjct: 181 QGVEAQTLANVYLALENNLEIIPVLNKIDLPGAEPEKVLREIEEVIGLDCSKAIFCSAKE 240
Query: 239 GVGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISS 298
G+G+ +L A+++RIP P + LR L+ DSY+D YRGVI + V+DG ++KGD+I
Sbjct: 241 GIGITEILDAIVQRIPAPLDTAGKPLRALIFDSYYDPYRGVIVYFRVIDGKVKKGDRIFF 300
Query: 299 AATGKSYEAMDIGIMHPELTPTGILFTGQVGYVITGMRTTKEARIGDTIYHTKSTVDVEP 358
A+GK Y A ++G++ P L+ G+VGY+ +R+ +AR+GDTI H +
Sbjct: 301 MASGKDYFADEVGVLSPNQIQVDELYAGEVGYIAASVRSVADARVGDTITHYSRKAE-SS 359
Query: 359 LPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDASVSVTKETSTALGLGFRCGFLG 418
LPG++ A MVF GLFP D F L A+EKL NDA++ ETS+A+G GFRCGFLG
Sbjct: 360 LPGYEEATPMVFCGLFPVDADQFPDLRDALEKLQLNDAALKFEPETSSAMGFGFRCGFLG 419
Query: 419 LLHMDVFHQRLEQEYGAHIISTVPTVPYIYEYSDGSKLEVQNPAALPSNPKQRVVACWEP 478
LLHM++ +RLE+EY ++I+T P+V Y +G NP+ LP +++ V EP
Sbjct: 420 LLHMEIVQERLEREYNLNLITTAPSVVYRVNSVNGDTTLCSNPSRLPDPGQRKSVE--EP 477
Query: 479 TVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELKSI 538
V ++ P +Y+G ++ L ERRGE E +I R + Y LPL E+V DF+++LKS
Sbjct: 478 YVKIELLTPKDYIGALMELAQERRGEFKEMKYIAENRASILYELPLAEMVGDFFDQLKSR 537
Query: 539 TSGYASFDYEDSDYQPSDLVKLDILLNGQAVDAMATIVHNSKSYRVGRELVEKLKKVIDR 598
T GYAS +Y Y+ SDL+KLDIL+N + V+ ++TIVH K+Y VGR L +KLK++I R
Sbjct: 538 TKGYASMEYSVIGYRESDLIKLDILINAEMVEPLSTIVHRDKAYSVGRALTQKLKELIPR 597
Query: 599 QMFEIIIQAAIGSKIIARETVTAMRKNVLAKCYGGDITRKKKLLEKQKEGKKRMKRVGSV 658
QMF++ IQA IGSK+IA E ++A+RK+VLAKCYGGDI+RKKKLL+KQ GKKRMK +G V
Sbjct: 598 QMFKVPIQACIGSKVIASEALSAIRKDVLAKCYGGDISRKKKLLKKQAAGKKRMKAIGRV 657
Query: 659 DVPQEAFHELLKV 671
DVPQEAF +LK+
Sbjct: 658 DVPQEAFMAVLKL 670
>At5g13650 GTP-binding protein typA (tyrosine phosphorylated protein
A)
Length = 609
Score = 148 bits (373), Expect = 1e-35
Identities = 140/524 (26%), Positives = 231/524 (43%), Gaps = 89/524 (16%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKG-LGQPQYLDKLQVERERGITVKAQTATMF 125
VRN +I+AHVDHGK+TL D +L + + Q + +D +ERERGIT+ ++ ++
Sbjct: 16 VRNIAIVAHVDHGKTTLVDSMLRQAKVFRDNQVMQERIMDSNDLERERGITILSKNTSIT 75
Query: 126 YKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQT 185
YKN +N+IDTPGH DF EV R L GVLLVVD+ +G QT
Sbjct: 76 YKNT---------------KVNIIDTPGHSDFGGEVERVLNMVDGVLLVVDSVEGPMPQT 120
Query: 186 VANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMF-DLDPSD------ALLTS--- 235
A E A++ V+NKID+P+A P+ V +F +L+ +D A+ S
Sbjct: 121 RFVLKKALEFGHAVVVVVNKIDRPSARPEFVVNSTFELFIELNATDEQCDFQAIYASGIK 180
Query: 236 AKTGVG-------LEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDG 288
K G+ L + A+I +P P + + +L+ML + +DE++G I + G
Sbjct: 181 GKAGLSPDDLAEDLGPLFEAIIRCVPGPNIEKDGALQMLATNIEYDEHKGRIAIGRLHAG 240
Query: 289 ALRKGDKISSAATGKSYEAMDIGIMHP----ELTPTGILFTGQVGYVITGMRTTKEARIG 344
LRKG + + S + + PT + G + + G+ +IG
Sbjct: 241 VLRKGMDVRVCTSEDSCRFARVSELFVYEKFYRVPTDSVEAGDI-CAVCGI---DNIQIG 296
Query: 345 DTIYHTKSTVDVEPLPGFKAAK---HMVFS----------GLFPADGSDFEALSHAIEKL 391
+TI V +PLP K + M FS G + + + L+ +E+
Sbjct: 297 ETI---ADKVHGKPLPTIKVEEPTVKMSFSVNTSPFSGREGKYVTSRNLRDRLNRELERN 353
Query: 392 TCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPYIYEYS 451
T +T G G LH+ + + + +E
Sbjct: 354 LAMKVEDGETADTFIVSG-------RGTLHITILIENMRRE------------------- 387
Query: 452 DGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFI 511
G + V P + +++ EP IAT+ +P ++GPV+ LL +RRG+ + +
Sbjct: 388 -GYEFMVGPPKVINKRVNDKLL---EPYEIATVEVPEAHMGPVVELLGKRRGQMFDMQGV 443
Query: 512 DSQ-RVFMKYRLPLREIVIDFYNELKSITSGYASFDYEDSDYQP 554
S+ F++Y++P R + + N + + + G A + Y P
Sbjct: 444 GSEGTTFLRYKIPTRGL-LGLRNAILTASRGTAILNTVFDSYGP 486
>At2g31060 putative GTP-binding protein
Length = 664
Score = 129 bits (324), Expect = 5e-30
Identities = 129/480 (26%), Positives = 202/480 (41%), Gaps = 78/480 (16%)
Query: 60 SQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKA 119
S P +RN ++IAHVDHGK+TL DRLL G + + +D + +ERERGIT+ +
Sbjct: 52 SSLDPNRLRNVAVIAHVDHGKTTLMDRLLRQCGA---DIPHERAMDSINLERERGITISS 108
Query: 120 QTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQ 179
+ ++F+K+ LN++DTPGH DF EV R + +G +LVVDA +
Sbjct: 109 KVTSIFWKD---------------NELNMVDTPGHADFGGEVERVVGMVEGAILVVDAGE 153
Query: 180 GVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMF--------DLDPSDA 231
G AQT A + L I ++NK+D+P+ D V+ + +F LD
Sbjct: 154 GPLAQTKFVLAKALKYGLRPILLLNKVDRPSERCDEVESLVFDLFANCGATEEQLD-FPV 212
Query: 232 LLTSAKTG--------------VGLEHVLPAVIERIPPPPGKSESSLRMLLLDSYFDEYR 277
L SAK G + +L AV+ + PP + ML+ D Y
Sbjct: 213 LYASAKEGWASSTYTKDPPVDAKNMADLLDAVVRHVQPPKANLDEPFLMLVSMMEKDFYL 272
Query: 278 GVICHVAVVDGALRKGDKI-----SSAATGKSYEAMDIGIMHPELTPTGILFTGQVGYVI 332
G I V G +R GD++ + + + K EA + +M + T + G +I
Sbjct: 273 GRILTGRVTSGVVRVGDRVNGLRKTDSGSEKIEEAKVVKLMKKKGTTIVSIDAAGAGDII 332
Query: 333 TGMRTTKEARIGDTIYHTK-----STVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHA 387
M IG T+ + TV+++P P + S L DG+
Sbjct: 333 C-MAGLTAPSIGHTVASAEVTTALPTVELDP-PTISMTFGVNDSPLAGQDGT--HLTGGR 388
Query: 388 IEKLTCNDASVSVTKETSTALGLGFRCGFLGLLHMDVFHQRLEQEYGAHIISTVPTVPYI 447
I +A ++ L + G L + + + + +E G + + P V +
Sbjct: 389 IGDRLMAEAETNLAINVIPGLSESYEVQGRGELQLGILIENMRRE-GFELSVSPPKV--M 445
Query: 448 YEYSDGSKLEVQNPAALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLE 507
Y+ G KL EP TI + E+VG V+ LS RR E ++
Sbjct: 446 YKTEKGQKL--------------------EPIEEVTIEINDEHVGLVMEALSHRRAEVID 485
>At1g56070 elongation factor like protein
Length = 843
Score = 105 bits (263), Expect = 6e-23
Identities = 58/144 (40%), Positives = 84/144 (58%), Gaps = 2/144 (1%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTIKKGL-GQPQYLDKLQVERERGITVKAQTATMF 125
+RN S+IAHVDHGKSTL D L+ G I + + G + D E ERGIT+K+ +++
Sbjct: 19 IRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEVAGDVRMTDTRADEAERGITIKSTGISLY 78
Query: 126 YKNIING-DDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQ 184
Y+ F ++ + YL+NLID+PGHVDFS EV+ +L G L+VVD +GV Q
Sbjct: 79 YEMTDESLKSFTGARDGNEYLINLIDSPGHVDFSSEVTAALRITDGALVVVDCIEGVCVQ 138
Query: 185 TVANFYLAFESNLAIIPVINKIDQ 208
T A + + +NK+D+
Sbjct: 139 TETVLRQALGERIRPVLTVNKMDR 162
>At3g22980 eukaryotic translation elongation factor 2, putative
Length = 1015
Score = 95.9 bits (237), Expect = 6e-20
Identities = 58/145 (40%), Positives = 83/145 (57%), Gaps = 18/145 (12%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTG---TIKKGLGQPQYLDKLQVERERGITVKAQTAT 123
VRN I+AHVDHGK+TLAD L+ +G + G+ +++D L E+ R IT+K+ + +
Sbjct: 9 VRNICILAHVDHGKTTLADHLIASSGGGVLHPRLAGKLRFMDYLDEEQRRAITMKSSSIS 68
Query: 124 MFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQA 183
+ YK+ Y LNLID+PGH+DF EVS + G L++VDA +GV
Sbjct: 69 LKYKD---------------YSLNLIDSPGHMDFCSEVSTAARLSDGALVLVDAVEGVHI 113
Query: 184 QTVANFYLAFESNLAIIPVINKIDQ 208
QT A A+ L V+NKID+
Sbjct: 114 QTHAVLRQAWIEKLTPCLVLNKIDR 138
Score = 31.2 bits (69), Expect = 2.0
Identities = 27/84 (32%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Query: 462 AALPSNPKQRVVACWEPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYR 521
A L +NP R+V E + EY+GP+ +LS RR L+ + +F +
Sbjct: 866 AVLQTNP--RIV---EAMYFCELNTAPEYLGPMYAVLSRRRARILKEEMQEGSSLFTVHA 920
Query: 522 -LPLREIVIDFYNELKSITSGYAS 544
+P+ E F +EL+ TSG AS
Sbjct: 921 YVPVSE-SFGFADELRKGTSGGAS 943
>At1g06220 elongation factor like protein
Length = 987
Score = 95.1 bits (235), Expect = 1e-19
Identities = 54/149 (36%), Positives = 83/149 (55%), Gaps = 14/149 (9%)
Query: 64 PELVRNFSIIAHVDHGKSTLADRLLELTGTIK----KGLGQPQYLDKLQVERERGITVKA 119
P LVRN +++ H+ HGK+ D L+E T + K +Y D E+ER I++KA
Sbjct: 135 PALVRNVALVGHLQHGKTVFMDMLVEQTHHMSTFNAKNEKHMKYTDTRVDEQERNISIKA 194
Query: 120 QTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQ 179
++ ++ S +YL N++DTPGHV+FS E++ SL G +L+VDAA+
Sbjct: 195 VPMSLVLED----------SRSKSYLCNIMDTPGHVNFSDEMTASLRLADGAVLIVDAAE 244
Query: 180 GVQAQTVANFYLAFESNLAIIPVINKIDQ 208
GV T A + +L I+ VINK+D+
Sbjct: 245 GVMVNTERAIRHAIQDHLPIVVVINKVDR 273
>At2g45030 putative mitochondrial translation elongation factor G
Length = 754
Score = 92.4 bits (228), Expect = 7e-19
Identities = 56/164 (34%), Positives = 89/164 (54%), Gaps = 25/164 (15%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTI--------KKGLGQPQYLDKLQVERERGITVK 118
+RN I AH+D GK+TL +R+L TG I + G+G +D + +ERE+GIT++
Sbjct: 65 LRNIGISAHIDSGKTTLTERVLFYTGRIHEIHEVRGRDGVGAK--MDSMDLEREKGITIQ 122
Query: 119 AQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAA 178
+ +K+ Y +N+IDTPGHVDF+ EV R+L G +LV+ +
Sbjct: 123 SAATYCTWKD---------------YKVNIIDTPGHVDFTIEVERALRVLDGAILVLCSV 167
Query: 179 QGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKS 222
GVQ+Q++ + + INK+D+ ADP +V Q ++
Sbjct: 168 GGVQSQSITVDRQMRRYEVPRVAFINKLDRMGADPWKVLNQARA 211
Score = 33.1 bits (74), Expect = 0.52
Identities = 36/159 (22%), Positives = 70/159 (43%), Gaps = 16/159 (10%)
Query: 278 GVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHP-ELTPTGILFTGQVGYVITGMR 336
G + ++ V +G ++KGD I + TGK + + MH ++ GQ+ V
Sbjct: 379 GQLTYLRVYEGVIKKGDFIINVNTGKRIKVPRLVRMHSNDMEDIQEAHAGQIVAVFGIEC 438
Query: 337 TTKEARIGDTIYHTKSTVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDA 396
+ + ++ +T ++++V P P A V G F S A+ + D
Sbjct: 439 ASGDTFTDGSVKYTMTSMNV-PEPVMSLAVQPVSKD----SGGQF---SKALNRFQKEDP 490
Query: 397 S--VSVTKETSTALGLGFRCGFLGLLHMDVFHQRLEQEY 433
+ V + E+ + G +G LH+D++ +R+ +EY
Sbjct: 491 TFRVGLDPESGQTIISG-----MGELHLDIYVERMRREY 524
>At1g45332 mitochondrial elongation factor, putative
Length = 754
Score = 92.4 bits (228), Expect = 7e-19
Identities = 56/164 (34%), Positives = 89/164 (54%), Gaps = 25/164 (15%)
Query: 67 VRNFSIIAHVDHGKSTLADRLLELTGTI--------KKGLGQPQYLDKLQVERERGITVK 118
+RN I AH+D GK+TL +R+L TG I + G+G +D + +ERE+GIT++
Sbjct: 65 LRNIGISAHIDSGKTTLTERVLFYTGRIHEIHEVRGRDGVGAK--MDSMDLEREKGITIQ 122
Query: 119 AQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAA 178
+ +K+ Y +N+IDTPGHVDF+ EV R+L G +LV+ +
Sbjct: 123 SAATYCTWKD---------------YKVNIIDTPGHVDFTIEVERALRVLDGAILVLCSV 167
Query: 179 QGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKS 222
GVQ+Q++ + + INK+D+ ADP +V Q ++
Sbjct: 168 GGVQSQSITVDRQMRRYEVPRVAFINKLDRMGADPWKVLNQARA 211
Score = 33.1 bits (74), Expect = 0.52
Identities = 36/159 (22%), Positives = 70/159 (43%), Gaps = 16/159 (10%)
Query: 278 GVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHP-ELTPTGILFTGQVGYVITGMR 336
G + ++ V +G ++KGD I + TGK + + MH ++ GQ+ V
Sbjct: 379 GQLTYLRVYEGVIKKGDFIINVNTGKRIKVPRLVRMHSNDMEDIQEAHAGQIVAVFGIEC 438
Query: 337 TTKEARIGDTIYHTKSTVDVEPLPGFKAAKHMVFSGLFPADGSDFEALSHAIEKLTCNDA 396
+ + ++ +T ++++V P P A V G F S A+ + D
Sbjct: 439 ASGDTFTDGSVKYTMTSMNV-PEPVMSLAVQPVSKD----SGGQF---SKALNRFQKEDP 490
Query: 397 S--VSVTKETSTALGLGFRCGFLGLLHMDVFHQRLEQEY 433
+ V + E+ + G +G LH+D++ +R+ +EY
Sbjct: 491 TFRVGLDPESGQTIISG-----MGELHLDIYVERMRREY 524
>At5g25230 translation Elongation Factor 2 - like protein
Length = 973
Score = 90.5 bits (223), Expect = 3e-18
Identities = 52/149 (34%), Positives = 82/149 (54%), Gaps = 14/149 (9%)
Query: 64 PELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQP----QYLDKLQVERERGITVKA 119
P LVRN +++ H+ HGK+ D L+E T + + +Y D E+ER I++KA
Sbjct: 121 PALVRNVALVGHLQHGKTVFMDMLVEQTHRMSTFNAENDKHMRYTDTRVDEQERNISIKA 180
Query: 120 QTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQ 179
++ ++ S +YL N++DTPG+V+FS E++ SL G + +VDAAQ
Sbjct: 181 VPMSLVLED----------SRSKSYLCNIMDTPGNVNFSDEMTASLRLADGAVFIVDAAQ 230
Query: 180 GVQAQTVANFYLAFESNLAIIPVINKIDQ 208
GV T A + +L I+ VINK+D+
Sbjct: 231 GVMVNTERAIRHAIQDHLPIVVVINKVDR 259
>At1g62750 elongation factor G, putative
Length = 783
Score = 82.0 bits (201), Expect = 1e-15
Identities = 55/190 (28%), Positives = 94/190 (48%), Gaps = 25/190 (13%)
Query: 68 RNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQY----LDKLQVERERGITVKAQTAT 123
RN I+AH+D GK+T +R+L TG K +G+ +D ++ E+ERGIT+ + T
Sbjct: 97 RNIGIMAHIDAGKTTTTERILYYTGRNYK-IGEVHEGTATMDWMEQEQERGITITSAATT 155
Query: 124 MFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQA 183
F+ + +N+IDTPGHVDF+ EV R+L G + + D+ GV+
Sbjct: 156 TFW---------------DKHRINIIDTPGHVDFTLEVERALRVLDGAICLFDSVAGVEP 200
Query: 184 QTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMFDLDPSDALLTSAKTGVGLE 243
Q+ + A + + I +NK+D+ A+ R + + + P + +G E
Sbjct: 201 QSETVWRQADKYGVPRICFVNKMDRLGANFFRTRDMIVTNLGAKP-----LVLQIPIGAE 255
Query: 244 HVLPAVIERI 253
V V++ +
Sbjct: 256 DVFKGVVDLV 265
Score = 32.3 bits (72), Expect = 0.88
Identities = 20/69 (28%), Positives = 35/69 (49%), Gaps = 1/69 (1%)
Query: 477 EPTVIATIVMPSEYVGPVITLLSERRGEQLEYSFIDSQRVFMKYRLPLREIVIDFYNELK 536
EP + +V P E++G VI L+ RRG+ + + +PL E + + + L+
Sbjct: 689 EPIMRVEVVTPEEHLGDVIGDLNSRRGQINSFGDKPGGLKVVDSLVPLAE-MFQYVSTLR 747
Query: 537 SITSGYASF 545
+T G AS+
Sbjct: 748 GMTKGRASY 756
>At4g11160 translation initiation factor IF-2 like protein
Length = 743
Score = 74.3 bits (181), Expect = 2e-13
Identities = 61/233 (26%), Positives = 105/233 (44%), Gaps = 35/233 (15%)
Query: 71 SIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYLDKLQVERERGITVKAQTATMFYKNII 130
+++ HVDHGK++L D L + ++ G Q++ V
Sbjct: 223 TVMGHVDHGKTSLLDALRNTSVAAREAGGITQHVGAFVV--------------------- 261
Query: 131 NGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFY 190
G S + +DTPGH FS +R A V+LVV A GV QT+
Sbjct: 262 -------GMPDSGTSITFLDTPGHAAFSEMRARGAAVTDIVVLVVAADDGVMPQTLEAIA 314
Query: 191 LAFESNLAIIPVINKIDQPTADPDRVKGQLKS----MFDLDPS-DALLTSAKTGVGLEHV 245
A +N+ ++ INK D+P A+P++VK QL S + D+ + A+ SA GL+ +
Sbjct: 315 HARSANVPVVVAINKCDKPGANPEKVKYQLTSEGIELEDIGGNVQAVEVSAAKSTGLDKL 374
Query: 246 LPAVIERIPPPPGKS--ESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKI 296
A++ + K+ + + ++++ D+ RG + + V G L +G +
Sbjct: 375 EEALLLQAVDMDLKARVDGPAQAYVVEARLDKGRGPLATIIVKAGTLVRGQHV 427
>At4g20360 translation elongation factor EF-Tu precursor,
chloroplast
Length = 476
Score = 67.4 bits (163), Expect = 2e-11
Identities = 83/370 (22%), Positives = 143/370 (38%), Gaps = 52/370 (14%)
Query: 35 FSITRALFCTQSRQNYTKEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTI 94
+S+T +R+++T A + P + N I HVDHGK+TL L +I
Sbjct: 49 YSLTTTSASQSTRRSFTVRAARGKFERKKPHV--NIGTIGHVDHGKTTLTAALTMALASI 106
Query: 95 KKGLGQP-QYLDKLQVERERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPG 153
+ + +D ER RGIT+ TAT+ Y E+ N +D PG
Sbjct: 107 GSSVAKKYDEIDAAPEERARGITIN--TATVEY-------------ETENRHYAHVDCPG 151
Query: 154 HVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVANFYLAFESNLA-IIPVINKIDQPTAD 212
H D+ + A G +LVV A G QT + LA + + ++ +NK DQ D
Sbjct: 152 HADYVKNMITGAAQMDGAILVVSGADGPMPQTKEHILLAKQVGVPDMVVFLNKEDQ-VDD 210
Query: 213 PDRVK------GQLKSMFDLDPSDALLTSAKTGVGLE--------------------HVL 246
+ ++ +L S ++ + D + S + +E ++
Sbjct: 211 AELLELVELEVRELLSSYEFNGDDIPIISGSALLAVETLTENPKVKRGDNKWVDKIYELM 270
Query: 247 PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYE 306
AV + IP P ++E + + D + RG + V G ++ G+ + ++
Sbjct: 271 DAVDDYIPIPQRQTELPFLLAVEDVFSITGRGTVATGRVERGTVKVGETVDLVGLRETRS 330
Query: 307 AMDIGIMHPELTPTGILFTGQVGYVITGM------RTTKEARIGDTIYHTKSTVDVEPLP 360
G+ + L VG ++ G+ R A+ G HTK + L
Sbjct: 331 YTVTGVEMFQKILDEALAGDNVGLLLRGIQKADIQRGMVLAKPGSITPHTKFEAIIYVLK 390
Query: 361 GFKAAKHMVF 370
+ +H F
Sbjct: 391 KEEGGRHSPF 400
>At1g17220 putative translation initiation factor IF2
Length = 1026
Score = 63.9 bits (154), Expect = 3e-10
Identities = 69/260 (26%), Positives = 110/260 (41%), Gaps = 40/260 (15%)
Query: 45 QSRQNYTKEKAIIDLSQYPPELVRNFSIIAHVDHGKSTLADRLLELTGTIKKGLGQPQYL 104
+ RQ + +E + L PP + +I+ HVDHGK+TL D + +
Sbjct: 484 KKRQTFDEED-LDKLEDRPPVI----TIMGHVDHGKTTLLDYIRK--------------- 523
Query: 105 DKLQVERERGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRS 164
K+ GIT YK + DGK S L DTPGH F +R
Sbjct: 524 SKVAASEAGGITQGIGA----YKVSVP----VDGKLQSCVFL---DTPGHEAFGAMRARG 572
Query: 165 LAACQGVLLVVDAAQGVQAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKSMF 224
++VV A G++ QT A + + I+ INKID+ A PDRV +L S+
Sbjct: 573 ARVTDIAIIVVAADDGIRPQTNEAIAHAKAAAVPIVIAINKIDKEGASPDRVMQELSSI- 631
Query: 225 DLDPSD------ALLTSAKTGVGLEHVLPAV--IERIPPPPGKSESSLRMLLLDSYFDEY 276
L P D + SA G ++ +L V + + + + +++++ D+
Sbjct: 632 GLMPEDWGGDVPMVQISALKGENVDDLLETVMLVAELQELKANPHRNAKGIVIEAGLDKA 691
Query: 277 RGVICHVAVVDGALRKGDKI 296
+G V G L++GD +
Sbjct: 692 KGPFATFIVQKGTLKRGDVV 711
>At5g10630 putative protein
Length = 804
Score = 58.9 bits (141), Expect = 9e-09
Identities = 57/184 (30%), Positives = 76/184 (40%), Gaps = 50/184 (27%)
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTIK-------------KGLGQPQY---LDKLQVERE 112
N +I+ HVD GKSTL+ RLL L G I +G G Y LD+ ERE
Sbjct: 378 NLAIVGHVDSGKSTLSGRLLHLLGRISQKQMHKYEKEAKLQGKGSFAYAWALDESAEERE 437
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ T + Y N S + + L+D+PGH DF + +
Sbjct: 438 RGITM---TVAVAYFN------------SKRHHVVLLDSPGHKDFVPNMIAGATQADAAI 482
Query: 173 LVVDAAQGV----------QAQTVANFYLAFESNLAIIPVINKIDQPTADPDRVKGQLKS 222
LV+DA+ G Q + A F I+ INK+D + G K
Sbjct: 483 LVIDASVGAFEAGFDNLKGQTREHARVLRGFGVEQVIV-AINKMD--------IVGYSKE 533
Query: 223 MFDL 226
FDL
Sbjct: 534 RFDL 537
>At5g60390 translation elongation factor eEF-1 alpha chain (gene A4)
Length = 449
Score = 58.5 bits (140), Expect = 1e-08
Identities = 72/296 (24%), Positives = 108/296 (36%), Gaps = 64/296 (21%)
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 69 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 113
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTA------------- 211
L++D+ G QT + LAF + +I NK+D T
Sbjct: 114 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVKQMICCCNKMDATTPKYSKARYDEIIKE 173
Query: 212 ----------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKSE 261
+PD++ S F+ D T+ G L +++I P S+
Sbjct: 174 VSSYLKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLLEALDQINEPKRPSD 231
Query: 262 SSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPEL 317
LR+ L D Y G + V G ++ G ++ A TG + E + + H L
Sbjct: 232 KPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTTEVKSVEMHHESL 287
>At1g07940 elongation factor 1-alpha
Length = 449
Score = 58.5 bits (140), Expect = 1e-08
Identities = 72/296 (24%), Positives = 108/296 (36%), Gaps = 64/296 (21%)
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 69 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 113
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTA------------- 211
L++D+ G QT + LAF + +I NK+D T
Sbjct: 114 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVKQMICCCNKMDATTPKYSKARYDEIIKE 173
Query: 212 ----------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKSE 261
+PD++ S F+ D T+ G L +++I P S+
Sbjct: 174 VSSYLKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLLEALDQINEPKRPSD 231
Query: 262 SSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPEL 317
LR+ L D Y G + V G ++ G ++ A TG + E + + H L
Sbjct: 232 KPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTTEVKSVEMHHESL 287
>At1g07930 elongation factor 1-alpha
Length = 449
Score = 58.5 bits (140), Expect = 1e-08
Identities = 72/296 (24%), Positives = 108/296 (36%), Gaps = 64/296 (21%)
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 69 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 113
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTA------------- 211
L++D+ G QT + LAF + +I NK+D T
Sbjct: 114 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVKQMICCCNKMDATTPKYSKARYDEIIKE 173
Query: 212 ----------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKSE 261
+PD++ S F+ D T+ G L +++I P S+
Sbjct: 174 VSSYLKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLLEALDQINEPKRPSD 231
Query: 262 SSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPEL 317
LR+ L D Y G + V G ++ G ++ A TG + E + + H L
Sbjct: 232 KPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTTEVKSVEMHHESL 287
>At1g07920 elongation factor 1-alpha
Length = 449
Score = 58.5 bits (140), Expect = 1e-08
Identities = 72/296 (24%), Positives = 108/296 (36%), Gaps = 64/296 (21%)
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTI----------------KKGLGQPQYLDKLQVERE 112
N +I HVD GKST L+ G I K+ LDKL+ ERE
Sbjct: 9 NIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVLDKLKAERE 68
Query: 113 RGITVKAQTATMFYKNIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVL 172
RGIT+ E++ Y +ID PGH DF + + +
Sbjct: 69 RGITIDIALWKF---------------ETTKYYCTVIDAPGHRDFIKNMITGTSQADCAV 113
Query: 173 LVVDAAQG-------VQAQTVANFYLAFESNL-AIIPVINKIDQPTA------------- 211
L++D+ G QT + LAF + +I NK+D T
Sbjct: 114 LIIDSTTGGFEAGISKDGQTREHALLAFTLGVKQMICCCNKMDATTPKYSKARYDEIIKE 173
Query: 212 ----------DPDRVKGQLKSMFDLDPSDALLTSAKTGVGLEHVLPAVIERIPPPPGKSE 261
+PD++ S F+ D T+ G L +++I P S+
Sbjct: 174 VSSYLKKVGYNPDKIPFVPISGFEGDNMIERSTNLDWYKG--PTLLEALDQINEPKRPSD 231
Query: 262 SSLRMLLLDSYFDEYRGVICHVAVVDGALRKGDKISSAATGKSYEAMDIGIMHPEL 317
LR+ L D Y G + V G ++ G ++ A TG + E + + H L
Sbjct: 232 KPLRLPLQDVYKIGGIGTVPVGRVETGMIKPGMVVTFAPTGLTTEVKSVEMHHESL 287
>At4g02930 mitochondrial elongation factor Tu
Length = 454
Score = 58.2 bits (139), Expect = 2e-08
Identities = 60/248 (24%), Positives = 103/248 (41%), Gaps = 36/248 (14%)
Query: 69 NFSIIAHVDHGKSTLADRLLELTGTIKKGLGQP-QYLDKLQVERERGITVKAQTATMFYK 127
N I HVDHGK+TL + ++ K +DK E++RGIT+ TA + Y
Sbjct: 69 NVGTIGHVDHGKTTLTAAITKVLAEEGKAKAIAFDEIDKAPEEKKRGITIA--TAHVEY- 125
Query: 128 NIINGDDFKDGKESSNYLLNLIDTPGHVDFSYEVSRSLAACQGVLLVVDAAQGVQAQTVA 187
E++ +D PGH D+ + A G +LVV G QT
Sbjct: 126 ------------ETAKRHYAHVDCPGHADYVKNMITGAAQMDGGILVVSGPDGPMPQTKE 173
Query: 188 NFYLAFESNL-AIIPVINKIDQPTADPDRVK------GQLKSMFDLDPSD---------A 231
+ LA + + +++ +NK+D DP+ ++ +L S + D +
Sbjct: 174 HILLARQVGVPSLVCFLNKVD-VVDDPELLELVEMELRELLSFYKFPGDDIPIIRGSALS 232
Query: 232 LLTSAKTGVGLEHVL---PAVIERIPPPPGKSESSLRMLLLDSYFDEYRGVICHVAVVDG 288
L +G + +L AV E IP P + M + D + + RG + + G
Sbjct: 233 ALQGTNDEIGRQAILKLMDAVDEYIPDPVRVLDKPFLMPIEDVFSIQGRGTVATGRIEQG 292
Query: 289 ALRKGDKI 296
++ G+++
Sbjct: 293 VIKVGEEV 300
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,339,431
Number of Sequences: 26719
Number of extensions: 609685
Number of successful extensions: 1792
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 1684
Number of HSP's gapped (non-prelim): 69
length of query: 672
length of database: 11,318,596
effective HSP length: 106
effective length of query: 566
effective length of database: 8,486,382
effective search space: 4803292212
effective search space used: 4803292212
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC139355.5


