

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139343.2 + phase: 0 /pseudo
(443 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
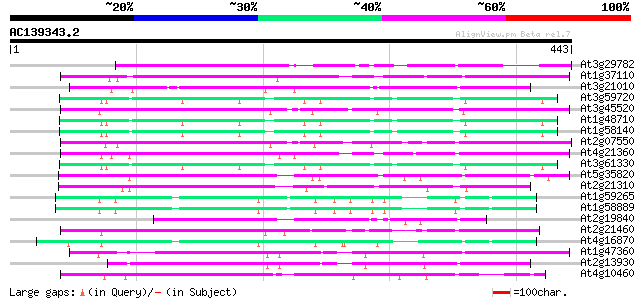
Score E
Sequences producing significant alignments: (bits) Value
At3g29782 hypothetical protein 164 9e-41
At1g37110 110 2e-24
At3g21010 unknown protein 99 5e-21
At3g59720 copia-type reverse transcriptase-like protein 97 2e-20
At3g45520 copia-like polyprotein 96 4e-20
At1g48710 hypothetical protein 96 5e-20
At1g58140 hypothetical protein 95 7e-20
At2g07550 putative retroelement pol polyprotein 94 2e-19
At4g21360 putative transposable element 92 7e-19
At3g61330 copia-type polyprotein 92 7e-19
At5g35820 copia-like retrotransposable element 86 3e-17
At2g21310 putative retroelement pol polyprotein 82 5e-16
At1g59265 polyprotein, putative 80 2e-15
At1g58889 polyprotein, putative 80 2e-15
At2g19840 copia-like retroelement pol polyprotein 80 2e-15
At2g21460 putative retroelement pol polyprotein 78 1e-14
At4g16870 retrotransposon like protein 76 3e-14
At1g47360 polyprotein, putative 76 4e-14
At2g13930 putative retroelement pol polyprotein 75 9e-14
At4g10460 putative retrotransposon 73 4e-13
>At3g29782 hypothetical protein
Length = 1130
Score = 164 bits (415), Expect = 9e-41
Identities = 106/361 (29%), Positives = 181/361 (49%), Gaps = 59/361 (16%)
Query: 84 IEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL 143
++ W + C+ IL+ + + LFD+Y K +K +W + KY +E+ Q+F +L
Sbjct: 117 MDTWAQGDYCCKGLILNRLLNDLFDLYSKAKSSKTLWLTLENKYKTDESRMQRFSTAKFL 176
Query: 144 *WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHK 203
++M + K I Q+ ++ +E++ E + + +VF L+EKL W D+K L K K
Sbjct: 177 NFKMVDSKPIMEQVEALQRICQEIELEGMSICNVFKTNCLIEKLPPGWSDFKNYLNFKRK 236
Query: 204 QMSLNDLIRHIIIEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPN 263
M+ +DLIR ++IE +R GA A+ Q N ++H K+
Sbjct: 237 AMTFDDLIRRLMIEGNNR---GA-------------HAGAQNQGHDVNVAEHKAKLKGKG 280
Query: 264 FKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHL 323
++ + + K GHKA C K + K +ANLT+ D + AV+++ ++
Sbjct: 281 KGFSIPQKNL--KIGHKADVC---KSKAKDVKSQANLTEED---------MVAVVTECNM 326
Query: 324 VNDVQ-KWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTS 382
V+D Q KW D+GAT HIC R F++Y++ +Q+++G++ + + GKGKV+LKLTS
Sbjct: 327 VDDNQVKWYYDTGATTHICTDRTMFSTYVN-NKSNEQLFMGNTAMSKIEGKGKVVLKLTS 385
Query: 383 GKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVL 442
G+ L L + SDK+V+ K+ V++G G+ GL +
Sbjct: 386 GRELTLQN---------------------------SDKLVLKKHGVYLGKGFVKGGLVKM 418
Query: 443 S 443
S
Sbjct: 419 S 419
>At1g37110
Length = 1356
Score = 110 bits (274), Expect = 2e-24
Identities = 108/433 (24%), Positives = 183/433 (41%), Gaps = 55/433 (12%)
Query: 41 IEVFKG-QNFRRWNERVFTLLDVHGVASVLTDVKPDEA----KSDAKQ------------ 83
I+VF G ++F W R+ L V G+ LTD + KS+AKQ
Sbjct: 10 IKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSGTKE 69
Query: 84 ------IEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKF 137
IE+ A + I++ ISD + Y D+W +N KY + +
Sbjct: 70 VPDPVKIEQSEQAKNI----IINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIY 125
Query: 138 VVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQ 197
++M I ++E+ +++ EL + I + + A ++ L +S K
Sbjct: 126 TQLKLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHT 185
Query: 198 LKHKHKQMSLNDL------IRHIIIEDTSRKECGAARAKALESRANLIQNNARKQQ-RYE 250
LK+ +K +++ D+ + + E + AA E L++NN + Q +
Sbjct: 186 LKYGNKTLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRPLVRNNQKGGQGKGR 245
Query: 251 NKSDHALKVTNPNFKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDD 310
++S+ KV C+ C K+GH C RK K + QG+ +
Sbjct: 246 SRSNSKTKVP----------CWYCKKEGHVKKDCYSRK------KKMESEGQGEAGVITE 289
Query: 311 DDVIAAVIS-KVHLVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTA 369
V + +S +V D+ W++DSG T H+ ++R+ F S+ G+ + LGD +
Sbjct: 290 KLVFSEALSVNEQMVKDL--WILDSGCTSHMTSRRDWFISFQEKGNTT--ILLGDDHSVE 345
Query: 370 VNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVF 429
G+G + + G L +V VP +R NLIS L+K+G + K+ KNN
Sbjct: 346 SQGQGTIRIDTHGGTIKILENVKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFKNNKT 405
Query: 430 VGNGYCDQGLFVL 442
G GL+VL
Sbjct: 406 ALRGSLSNGLYVL 418
>At3g21010 unknown protein
Length = 438
Score = 99.0 bits (245), Expect = 5e-21
Identities = 97/385 (25%), Positives = 169/385 (43%), Gaps = 27/385 (7%)
Query: 48 NFRRWNERVFTLLDVHGVASVLTD-VKPDEAKS-------DAKQIEKWNN-ANKVCR--H 96
N+ W T L G+ V+ + V PD +K A++ KW + A K +
Sbjct: 16 NYEIWAPIAKTTLTEKGLWDVVENGVPPDPSKVPELAATIQAEEFSKWRDLAEKDMKALQ 75
Query: 97 TILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIKVQ 156
+ S+++D+ F S AKD+WD++ K E+A K + + + M E + I
Sbjct: 76 ILQSSLTDSAFRKTLSASSAKDLWDSLE-KGNNEQA-KLRRLEKQFEELSMNEGEPINPY 133
Query: 157 INEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKH--KHKQMSLNDLIRHI 214
I+ +++E+L+ I D V ++ L S+DD L K MSL +
Sbjct: 134 IDRVIEIVEQLRRFKIAKSDYQVIEKVLATLSGSYDDVAPVLGDLVDLKNMSLKSFVELF 193
Query: 215 IIEDTSRKE-------CGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPN-FKA 266
+ D+ +E R+ A E L N KQ+ + A K + K
Sbjct: 194 YVYDSITEESIHLMLKANRLRSMAEEESCVLCNKNNHKQEDCSSSIPKAGKSSGARQSKP 253
Query: 267 NLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVND 326
G+C+ CG++GHKA C +K+ QP + + ++K+ + D ++ A ++ D
Sbjct: 254 KKGKCFQCGERGHKAKDCN-KKIQ-QPAVTEDQIEIVEEKEIEVDFLLLAALNLNEFTYD 311
Query: 327 VQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTL 386
W++ AT H+ + FT+ + +V L D V G G V +K+ GK
Sbjct: 312 ENMWMIYGHATNHMTPYEKLFTTLDRT--YKAKVGLVDGTIIMVEGIGDVKIKMKEGKQK 369
Query: 387 ALSDVLNVPTIRTNLISVALLNKVG 411
+++V+ VP + N++SV + G
Sbjct: 370 TINNVIFVPGLNRNVLSVQQMKSRG 394
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 96.7 bits (239), Expect = 2e-20
Identities = 99/458 (21%), Positives = 185/458 (39%), Gaps = 83/458 (18%)
Query: 40 RIEVFKGQNFRRWNERVFTLLDVHGVASVLTD--VKPD------EAKSDAKQIEKWNNAN 91
++ V N+ W+ R+ +L H V ++ ++P+ + + D + + +
Sbjct: 9 QVPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKK 68
Query: 92 KVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQ---KFVVGNYL*WQMK 148
+C I + + F+ AK+ W+ + Y + K+ + + G + QMK
Sbjct: 69 ALC--LIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMK 126
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVA-------------------------AAL 183
E + + + + LK L DV + A
Sbjct: 127 EGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMT 186
Query: 184 VEKLRSSWDDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKA----LESRANLI 239
+E+L S Y+++ K K D++ ++ +++E G + + + R
Sbjct: 187 IEQLLGSLQAYEEKKKKKE------DIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGG 240
Query: 240 QNNAR--------KQQRYENKSDHALKV-TNPNFKANLGECYVCGKKGHKAYHCRYRKVN 290
N R QR EN S K + + +CY CGK GH A C+ +
Sbjct: 241 YGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECK--APS 298
Query: 291 GQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSY 350
+ K KAN + +K ++D++ K + KW +DSGA+ H+C ++ F
Sbjct: 299 NKKFKEKANYVE---EKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAEL 355
Query: 351 ISVGDDED---QVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALL 407
DE V LGD V GKG ++++L +G +S+V +P+++TN++S+ L
Sbjct: 356 -----DESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
Query: 408 NKVGVKVSFESD-------------KMVMSKNNVFVGN 432
+ G + + + K+ MSKN +FV N
Sbjct: 411 LEKGYDIRLKDNNLSIRDKESNLITKVPMSKNRMFVLN 448
>At3g45520 copia-like polyprotein
Length = 1363
Score = 95.9 bits (237), Expect = 4e-20
Identities = 88/430 (20%), Positives = 190/430 (43%), Gaps = 39/430 (9%)
Query: 41 IEVFKGQ-NFRRWNERVFTLLDVHGVASVL------------TDVKPDEAKSDAKQIEKW 87
+E F G+ ++ W E++ +D+ G+++VL ++ ++ K + +++E +
Sbjct: 8 VEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEEREKMEAF 67
Query: 88 NNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQM 147
+ R TI+ ++SD + A + + ++ Y ++ + ++ ++M
Sbjct: 68 EEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKLYSFKM 127
Query: 148 KEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQ--M 205
E+ I+ I+E+ ++ +L+ N+++ D A L+ L +D K LK+ + +
Sbjct: 128 SENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSSGKTVL 187
Query: 206 SLNDLIRHIIIEDTSRKECGAARAKALESRAN--LIQNNARKQQRYENKSDHALKVTNPN 263
SL+++ I + E G+ + K+++ +A +++ A + R E K K +
Sbjct: 188 SLDEVAAAIYSREL---EFGSVK-KSIKGQAEGLYVKDKAENRGRSEQKDKGKGKRSKSK 243
Query: 264 FKANLGECYVCGKKGHKAYHCRYRKV---------NGQPPKPKANLTQGDDKKYDDDDVI 314
K C++CG+ GH C + G+ K NL +G + +
Sbjct: 244 SKRG---CWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVNFVESAGMF 300
Query: 315 AAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSYISVGDDE--DQVYLGDSRTTAVNG 372
+ ++ +W++D+G H+ KRE + D+E V +G+ + V G
Sbjct: 301 VSEALSSTDIHLEDEWIMDTGCIYHMTHKREWLEDF----DEEAGGSVRMGNKSISRVKG 356
Query: 373 KGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGN 432
G V + +G T+ L +V +P + NL+S+ K G K E+ + + N +
Sbjct: 357 VGTVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGHKFESENGMLRIKSGNQVLLE 416
Query: 433 GYCDQGLFVL 442
G L++L
Sbjct: 417 GRRYDTLYIL 426
>At1g48710 hypothetical protein
Length = 1352
Score = 95.5 bits (236), Expect = 5e-20
Identities = 99/458 (21%), Positives = 185/458 (39%), Gaps = 83/458 (18%)
Query: 40 RIEVFKGQNFRRWNERVFTLLDVHGVASVLTD--VKPD------EAKSDAKQIEKWNNAN 91
++ V N+ W+ R+ +L H V ++ ++P+ + + D + + +
Sbjct: 9 QVPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKK 68
Query: 92 KVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQ---KFVVGNYL*WQMK 148
+C I + + F+ AK+ W+ + Y + K+ + + G + QMK
Sbjct: 69 ALC--LIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMK 126
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVA-------------------------AAL 183
E + + + + LK L DV + A
Sbjct: 127 EGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMT 186
Query: 184 VEKLRSSWDDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKA----LESRANLI 239
+E+L S Y+++ K K D+I ++ +++E G + + + R
Sbjct: 187 IEQLLGSLQAYEEKKKKKE------DIIEQVLNMQITKEENGQSYQRRGGGQVRGRGRGG 240
Query: 240 QNNAR--------KQQRYENKSDHALKV-TNPNFKANLGECYVCGKKGHKAYHCRYRKVN 290
N R QR EN S K + + +CY CGK GH A C+ +
Sbjct: 241 YGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECK--APS 298
Query: 291 GQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSY 350
+ + KAN + +K ++D++ K + KW +DSGA+ H+C ++ F
Sbjct: 299 NKKFEEKANYVE---EKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAEL 355
Query: 351 ISVGDDED---QVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALL 407
DE V LGD V GKG ++++L +G +S+V +P+++TN++S+ L
Sbjct: 356 -----DESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
Query: 408 NKVGVKVSFESD-------------KMVMSKNNVFVGN 432
+ G + + + K+ MSKN +FV N
Sbjct: 411 LEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLN 448
>At1g58140 hypothetical protein
Length = 1320
Score = 95.1 bits (235), Expect = 7e-20
Identities = 98/458 (21%), Positives = 185/458 (39%), Gaps = 83/458 (18%)
Query: 40 RIEVFKGQNFRRWNERVFTLLDVHGVASVLTD--VKPD------EAKSDAKQIEKWNNAN 91
++ V N+ W+ R+ +L H V ++ ++P+ + + D + + +
Sbjct: 9 QVPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKK 68
Query: 92 KVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQ---KFVVGNYL*WQMK 148
+C I + + F+ AK+ W+ + Y + K+ + + G + QMK
Sbjct: 69 ALC--LIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMK 126
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVA-------------------------AAL 183
E + + + + LK L DV + A
Sbjct: 127 EGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMT 186
Query: 184 VEKLRSSWDDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKA----LESRANLI 239
+E+L S Y+++ K K D++ ++ +++E G + + + R
Sbjct: 187 IEQLLGSLQAYEEKKKKKE------DIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGG 240
Query: 240 QNNAR--------KQQRYENKSDHALKV-TNPNFKANLGECYVCGKKGHKAYHCRYRKVN 290
N R QR EN S K + + +CY CGK GH A C+ +
Sbjct: 241 YGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECK--APS 298
Query: 291 GQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSY 350
+ + KAN + +K ++D++ K + KW +DSGA+ H+C ++ F
Sbjct: 299 NKKFEEKANYVE---EKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAEL 355
Query: 351 ISVGDDED---QVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALL 407
DE V LGD V GKG ++++L +G +S+V +P+++TN++S+ L
Sbjct: 356 -----DESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
Query: 408 NKVGVKVSFESD-------------KMVMSKNNVFVGN 432
+ G + + + K+ MSKN +FV N
Sbjct: 411 LEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLN 448
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 93.6 bits (231), Expect = 2e-19
Identities = 91/433 (21%), Positives = 191/433 (44%), Gaps = 50/433 (11%)
Query: 41 IEVFKGQ-NFRRWNERVFTLLDVHGVASVLTDVKP--------DEAKSDAKQ----IEKW 87
+E F G+ ++ W E++ +D+ G+ + L + + DE+ D ++ E
Sbjct: 8 VEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEAL 67
Query: 88 NNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQM 147
K R I+ +++D + A + ++ Y ++ + + ++M
Sbjct: 68 EEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFKM 127
Query: 148 KEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQ--M 205
E+ ++ I+E+ Q+I +L+ N+I+ D A L+ L ++D K LK+ + +
Sbjct: 128 SENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSIL 187
Query: 206 SLNDLIRHIIIEDTSRKECGAARAKALESRANLI------QNNARKQQRYENKSDHALKV 259
+L+++ I ++ E G+ + K+++ +A + +N + +Q+ + K
Sbjct: 188 TLDEVAAAIYSKEL---ELGSVK-KSIKVQAEGLYVKDKNENKGKGEQKGKGKGKKGKSK 243
Query: 260 TNPNFKANLGECYVCGKKGHKAYHC---------RYRKVNGQPPKPKANLTQGDDKKYDD 310
P C+ CG++GH C + + V G+ K NL + +
Sbjct: 244 KKPG-------CWTCGEEGHFRSSCPNQNKPQFKQSQVVKGESSGGKGNLAEAAGYYVSE 296
Query: 311 DDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAV 370
A ++VHL ++ W++D+G + H+ KRE F + D V +G+ + V
Sbjct: 297 ----ALSSTEVHLEDE---WILDTGCSYHMTYKREWFHEFNE--DAGGSVRMGNKTVSRV 347
Query: 371 NGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFV 430
G G + +K + G T+ L++V +P + NL+S+ K G K E + + N +
Sbjct: 348 RGVGTIRVKNSDGLTIVLTNVRYIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVL 407
Query: 431 GNGYCDQGLFVLS 443
G L++L+
Sbjct: 408 LTGRRYDTLYLLN 420
>At4g21360 putative transposable element
Length = 1308
Score = 91.7 bits (226), Expect = 7e-19
Identities = 99/430 (23%), Positives = 181/430 (42%), Gaps = 47/430 (10%)
Query: 41 IEVFKG-QNFRRWNERVFTLLDVHGVASVLTD---------VKPDEAKS-------DAKQ 83
I+ F G ++F W R+ L V G+ L+D VK ++ +S D+K+
Sbjct: 9 IKTFNGDRDFSLWKIRIEAQLGVLGLKPALSDFTLTKTILVVKSEKKESESEDDETDSKK 68
Query: 84 IEKWNNANKV-----CRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFV 138
E+ + K ++ I++ I+D + A ++W +N + + +
Sbjct: 69 TEEVPDPIKFEQSDQAKNFIINHITDTVLLKVQHCVTAAELWATLNKLFMETSLPNRIYT 128
Query: 139 VGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQL 198
++M ++ I +E+ +++ EL + I + + A ++ L S+ K L
Sbjct: 129 QLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAILILNSLPPSYIQLKHTL 188
Query: 199 KHKHKQMSLNDLI---RHIIIEDTSRKE---CGAARAKALESRANLIQNNARKQQRYENK 252
K+ +K +S+ D++ + + E + +KE A+ A R N + Q + +
Sbjct: 189 KYGNKTLSVQDVVSSAKSLERELSEQKETIRAPASTALYTAERGRPQTKNTQGQGKGRGR 248
Query: 253 SDHALKVTNPNFKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDD 312
S+ ++T C+ C K+GH C K + N QG +
Sbjct: 249 SNSKSRLT----------CWFCKKEGHVKKDCYAGK------RKLENEGQGKAGVITEKL 292
Query: 313 VIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNG 372
V + +S ++ KWV+DSG T H+ ++ + F+ + ++ + LGD T G
Sbjct: 293 VYSEALS-MYDQEAKDKWVIDSGCTYHMTSRMDWFSEFNE--NETTMILLGDDHTVESKG 349
Query: 373 KGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGN 432
G V + G L +V VP +R NLIS L+K+G K K+ K N
Sbjct: 350 SGTVKVNTHGGSIRVLKNVRFVPNLRRNLISTGTLDKLGYKHEGGDGKVRFYKENKTALC 409
Query: 433 GYCDQGLFVL 442
G GL+VL
Sbjct: 410 GNLVNGLYVL 419
>At3g61330 copia-type polyprotein
Length = 1352
Score = 91.7 bits (226), Expect = 7e-19
Identities = 97/458 (21%), Positives = 184/458 (39%), Gaps = 83/458 (18%)
Query: 40 RIEVFKGQNFRRWNERVFTLLDVHGVASVLTD--VKPD------EAKSDAKQIEKWNNAN 91
++ V N+ W+ R+ +L H V ++ ++P+ + + D + + +
Sbjct: 9 QVPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKK 68
Query: 92 KVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQ---KFVVGNYL*WQMK 148
+C I + + F+ AK+ W+ + Y + K+ + + G + QMK
Sbjct: 69 ALC--LIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMK 126
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVA-------------------------AAL 183
E + + + + LK L DV + A
Sbjct: 127 EGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMT 186
Query: 184 VEKLRSSWDDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKA----LESRANLI 239
+E+L S Y+++ K K D+ ++ +++E G + + + R
Sbjct: 187 IEQLLGSLQAYEEKKKKKE------DIAEQVLNMQITKEENGQSYQRRGGGQVRGRGRGG 240
Query: 240 QNNAR--------KQQRYENKSDHALKV-TNPNFKANLGECYVCGKKGHKAYHCRYRKVN 290
N R QR EN S K + + +CY CGK GH A C+ +
Sbjct: 241 YGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECK--APS 298
Query: 291 GQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSY 350
+ + KA+ + +K ++D++ K + KW +DSGA+ H+C ++ F
Sbjct: 299 NKKFEEKAHYVE---EKIQEEDMLLMASYKKDEQKENHKWYLDSGASNHMCGRKSMFAEL 355
Query: 351 ISVGDDED---QVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALL 407
DE V LGD V GKG ++++L +G +S+V +P+++TN++S+ L
Sbjct: 356 -----DESVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
Query: 408 NKVGVKVSFESD-------------KMVMSKNNVFVGN 432
+ G + + + K+ MSKN +FV N
Sbjct: 411 LEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLN 448
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 86.3 bits (212), Expect = 3e-17
Identities = 92/437 (21%), Positives = 186/437 (42%), Gaps = 53/437 (12%)
Query: 39 SRIEVFKGQ-NFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKV---- 93
+ +E F G ++ W E++ +++ G+ L + + + +I N +
Sbjct: 6 AEVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATS 65
Query: 94 -------------CRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVG 140
R TI+ ++ + + K A + ++ + A+ + ++
Sbjct: 66 KLEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQ 125
Query: 141 NYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKH 200
++M E+ ++ +N++ +LI +L+ +++PD A L+ L +D K+ LK+
Sbjct: 126 RLYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKY 185
Query: 201 KHKQMSLNDLIRHIIIEDTSRKECGAARAKALESRAN--LIQNNA-----RKQQRYENKS 253
+ L ++ A R+K LE A+ L++NN+ + + R E +
Sbjct: 186 CKTTLHLEEIT-------------SAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRG 232
Query: 254 DHALKVTNPNFKANLGE-CYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDD-- 310
K + + G+ C++CGK+GH C K K + +G+
Sbjct: 233 KGPNKNKSRSKSKGAGKTCWICGKEGHFKKQCYVWKERN---KQGSTSERGEASTVTARV 289
Query: 311 DDVIAAVISKVHL---VNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRT 367
D A V+S+ L W++D+G + H+ +++ + + V +G+
Sbjct: 290 TDAAALVVSRALLGFAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGK--VRMGNDTY 347
Query: 368 TAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMS--K 425
+ V G G V +K G T+ L+DV +P + NLIS+ L G FES K +++ K
Sbjct: 348 SEVKGIGDVRIKNEDGSTILLTDVRYIPEMSKNLISLGTLEDKG--CWFESKKGILTIFK 405
Query: 426 NNVFVGNGYCDQGLFVL 442
N++ V G + L+ L
Sbjct: 406 NDLTVLTGKKESTLYFL 422
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 82.4 bits (202), Expect = 5e-16
Identities = 85/402 (21%), Positives = 171/402 (42%), Gaps = 52/402 (12%)
Query: 39 SRIEVFKGQ-NFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKW--NNANKV-- 93
S +E G+ ++ W E++ +++ G+ L + + E + + + +KV
Sbjct: 6 SEVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLK 65
Query: 94 -----CRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMK 148
R T++ ++ + + K A + ++ + A+ + ++ ++M
Sbjct: 66 EKRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMS 125
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLN 208
+ I+ +N++ +LI +L+ + +PD A L+ L +D K LK+ ++L+
Sbjct: 126 DSMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKTTLALD 185
Query: 209 DLIRHIIIEDTSRKECGAARAKALE-----------SRANLIQNNARKQQRYENKSDHAL 257
++ GA R+K LE S A +Q+ R ++R +KS
Sbjct: 186 EI-------------TGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKR--DKSSERN 230
Query: 258 KVTNPNFKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAV 317
K + + C+VCGK+GH C K + K N +G+ AA
Sbjct: 231 KSQSRSKSREKKVCWVCGKEGHFKKQCYVWK---EKNKKGNNSEKGESSNVIGQAADAAA 287
Query: 318 ISKVHLVN-DVQ----KWVVDSGATRHICAKREAFTSYISVGDDEDQ---VYLGDSRTTA 369
++ N D Q +W++D+G + H+ +R+ F + DE Q V + + +
Sbjct: 288 LAVREESNADNQEVDNEWIMDTGCSFHMTPRRDWFVEF-----DESQTGRVKMANQTYSE 342
Query: 370 VNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVG 411
+ G G + ++ T+ L +V VP++ NLIS+ L G
Sbjct: 343 IKGIGSIRIQNDDNTTVLLKNVRYVPSMSKNLISMGTLEDQG 384
>At1g59265 polyprotein, putative
Length = 1466
Score = 80.5 bits (197), Expect = 2e-15
Identities = 100/428 (23%), Positives = 170/428 (39%), Gaps = 78/428 (18%)
Query: 37 DVSRIEVFKGQNFRRWNERVFTLLDVHGVASVL---TDVKPDEAKSDAK-----QIEKWN 88
++S + N+ W+ +V L D + +A L T + P +DA +W
Sbjct: 19 NMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTDAAPRVNPDYTRWK 78
Query: 89 NANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMK 148
+K+ +L IS ++ A IW+ + Y + L Q+K
Sbjct: 79 RQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYA-----NPSYGHVTQLRTQLK 133
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEK-LRSSWDDYK---QQLKHKHKQ 204
+ + I++Y Q + + +L VE+ L + ++YK Q+ K
Sbjct: 134 QWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAAKDTP 193
Query: 205 MSLNDLIRHIIIEDTSRKECGAARAKALESRANLIQ----------NNARKQQRYENKSD 254
+L ++ ++ ++ K + A + AN + NN + RY+N+++
Sbjct: 194 PTLTEIHERLLNHES--KILAVSSATVIPITANAVSHRNTTTTNNNNNGNRNNRYDNRNN 251
Query: 255 H----ALKVTNPNFKAN-------LGECYVCGKKGHKAYHCR-----YRKVNGQPP---- 294
+ + ++ NF N LG+C +CG +GH A C VN Q P
Sbjct: 252 NNNSKPWQQSSTNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQPPSPF 311
Query: 295 ---KPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSY- 350
+P+ANL G ++ W++DSGAT HI + + +
Sbjct: 312 TPWQPRANLALGSPYSSNN-------------------WLLDSGATHHITSDFNNLSLHQ 352
Query: 351 -ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISV-ALLN 408
+ GDD V + D T ++ G L T + L L ++L VP I NLISV L N
Sbjct: 353 PYTGGDD---VMVADGSTIPISHTGSTSLS-TKSRPLNLHNILYVPNIHKNLISVYRLCN 408
Query: 409 KVGVKVSF 416
GV V F
Sbjct: 409 ANGVSVEF 416
>At1g58889 polyprotein, putative
Length = 1466
Score = 80.5 bits (197), Expect = 2e-15
Identities = 100/428 (23%), Positives = 170/428 (39%), Gaps = 78/428 (18%)
Query: 37 DVSRIEVFKGQNFRRWNERVFTLLDVHGVASVL---TDVKPDEAKSDAK-----QIEKWN 88
++S + N+ W+ +V L D + +A L T + P +DA +W
Sbjct: 19 NMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTDAAPRVNPDYTRWK 78
Query: 89 NANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMK 148
+K+ +L IS ++ A IW+ + Y + L Q+K
Sbjct: 79 RQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYA-----NPSYGHVTQLRTQLK 133
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEK-LRSSWDDYK---QQLKHKHKQ 204
+ + I++Y Q + + +L VE+ L + ++YK Q+ K
Sbjct: 134 QWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAAKDTP 193
Query: 205 MSLNDLIRHIIIEDTSRKECGAARAKALESRANLIQ----------NNARKQQRYENKSD 254
+L ++ ++ ++ K + A + AN + NN + RY+N+++
Sbjct: 194 PTLTEIHERLLNHES--KILAVSSATVIPITANAVSHRNTTTTNNNNNGNRNNRYDNRNN 251
Query: 255 H----ALKVTNPNFKAN-------LGECYVCGKKGHKAYHCR-----YRKVNGQPP---- 294
+ + ++ NF N LG+C +CG +GH A C VN Q P
Sbjct: 252 NNNSKPWQQSSTNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQPPSPF 311
Query: 295 ---KPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSY- 350
+P+ANL G ++ W++DSGAT HI + + +
Sbjct: 312 TPWQPRANLALGSPYSSNN-------------------WLLDSGATHHITSDFNNLSLHQ 352
Query: 351 -ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISV-ALLN 408
+ GDD V + D T ++ G L T + L L ++L VP I NLISV L N
Sbjct: 353 PYTGGDD---VMVADGSTIPISHTGSTSLS-TKSRPLNLHNILYVPNIHKNLISVYRLCN 408
Query: 409 KVGVKVSF 416
GV V F
Sbjct: 409 ANGVSVEF 416
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 80.1 bits (196), Expect = 2e-15
Identities = 62/268 (23%), Positives = 124/268 (46%), Gaps = 26/268 (9%)
Query: 114 KVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENII 173
K A + W ++ Y + + ++ ++M++ K ++ ++E+ ++I +L I
Sbjct: 64 KTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYRMQDSKTLEENVDEFQKMISDLNNLQIQ 123
Query: 174 LPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKALE 233
+PD A ++ L S+D K+ LK+ + + L+D+I AA++K LE
Sbjct: 124 VPDEVQAILILSALPDSYDMLKETLKYGREGIKLDDVI-------------SAAKSKELE 170
Query: 234 SRANLIQNNARKQQRYENKSDHALKVTNPNFKANLGECYVCGKKGHKAYHCRYRKVNGQP 293
R + + + Y A P C++CGK+GH C Y+ +
Sbjct: 171 LRDSSGGSRPVGEGLYVRGKSQARGSDGPKSTEGKKVCWICGKEGHFKRQC-YKWLE--- 226
Query: 294 PKPKANLTQGDDKKYDDD--DVIAAVISKVHL---VNDVQKWVVDSGATRHICAKREAFT 348
K KAN G+ DD D++ V S+V++ +D ++W++D+G + H+ ++E
Sbjct: 227 -KNKAN-GAGETALVKDDAQDLVGLVASEVNMSEGKDDQEEWIMDTGCSFHMTPRKEYLM 284
Query: 349 SYISVGDDEDQVYLGDSRTTAVNGKGKV 376
++ +V + ++ + V G GKV
Sbjct: 285 DFVEA--KSGKVRMANNSFSEVKGIGKV 310
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 77.8 bits (190), Expect = 1e-14
Identities = 80/396 (20%), Positives = 170/396 (42%), Gaps = 48/396 (12%)
Query: 41 IEVFKGQ-NFRRWNERVFTLLDVHGVASVLTD------------VKPDEAKSDAKQIEKW 87
+E F G+ ++ W E++ LD+ G++ L + + +E K + + E
Sbjct: 8 VEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRRELL 67
Query: 88 NNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQM 147
+ R I+ +++D + + A + ++ Y ++ + + ++M
Sbjct: 68 EEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFKM 127
Query: 148 KEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKH--KHKQM 205
E+ I+ I+E+ ++I +L+ N+++ D A L+ L +D + LK+ +
Sbjct: 128 SENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVTL 187
Query: 206 SLNDLIRHII---IEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNP 262
SL++++ I +E S K+ +A+ L + + R +QR N ++ + +
Sbjct: 188 SLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEK-TETRGRTEQRGNNNNNKKSRSKSR 246
Query: 263 NFKANLGECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVH 322
+ K C++CG+ + + Y + NG + T +H
Sbjct: 247 SKKG----CWICGESSNGS--SNYSEANGLYVSEALSSTD------------------IH 282
Query: 323 LVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTS 382
L ++ WV+D+G + H+ KRE F D V +G+ + V G G + +K +
Sbjct: 283 LEDE---WVMDTGCSYHMTYKREWFEDLNE--DAGGSVRMGNKTVSKVRGIGTIRVKNEA 337
Query: 383 GKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFES 418
G + L++V +P + NL+S+ K G E+
Sbjct: 338 GMVVRLTNVRYIPEMDRNLLSLGTFEKSGYSFKLEN 373
>At4g16870 retrotransposon like protein
Length = 1474
Score = 76.3 bits (186), Expect = 3e-14
Identities = 99/435 (22%), Positives = 164/435 (36%), Gaps = 67/435 (15%)
Query: 22 ATNPPAVVTYAKQFPDVSRIEVFK--GQNFRRWNERVFTLLDVHGVASVLTD-------- 71
AT A+V + +++ V K N+ W+ ++ LLD + +A L
Sbjct: 10 ATTDEAIVFTPQTIFNINTSNVTKLTSNNYLMWSLQIHALLDGYELAGHLDGSIETPAPT 69
Query: 72 VKPDEAKSDAKQIEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEE 131
+ + S Q W +++ ++ IS + + A IW + Y
Sbjct: 70 LTTNNVVSANPQYTLWKRQDRLIFSALIGAISPPVQPLVSRATKASQIWKTLTNTYA--- 126
Query: 132 ATKQKFVVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSW 191
K + L Q+K+ K+ I+EY L + IL VE++
Sbjct: 127 --KSSYDHIKQLRTQIKQLKKGTKTIDEYVLSHTTLLDQLAILGKPMEHEEQVERILEGL 184
Query: 192 -DDYK---QQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKALESRANLIQ----NNA 243
+DYK Q++ K S+ ++ +I + A + +L AN+ Q NN
Sbjct: 185 PEDYKTVVDQIEGKDNTPSITEIHERLINHEAKLLSTAALSSSSLPMSANVAQQRHHNNN 244
Query: 244 RKQQRYENKSDHALKVTN--PN---------FKANLGECYVCGKKGHKAYHCRYRKV--- 289
R + +N++ N P+ FK LG+C +C +GH A C +
Sbjct: 245 RNNNQNKNRTQGNTYTNNWQPSANNKSGQRPFKPYLGKCQICNVQGHSARRCPQLQAMQP 304
Query: 290 -------NGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICA 342
P +P+ANL G W++DSGAT HI +
Sbjct: 305 SSSSSASTFTPWQPRANLAMG-------------------APYTANNWLLDSGATHHITS 345
Query: 343 KREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLI 402
A + D+ V + D + + G L ++ + L L+ VL VP I+ NL+
Sbjct: 346 DLNALALHQPYNGDD--VMIADGTSLKITKTGSTFLP-SNARDLTLNKVLYVPDIQKNLV 402
Query: 403 SV-ALLNKVGVKVSF 416
SV L N V V F
Sbjct: 403 SVYRLCNTNQVSVEF 417
>At1g47360 polyprotein, putative
Length = 1182
Score = 75.9 bits (185), Expect = 4e-14
Identities = 80/412 (19%), Positives = 168/412 (40%), Gaps = 57/412 (13%)
Query: 48 NFRRWNERVFTLLDVHGVASV-----LTDVKPDEAKSDAKQIEKWNNANKVCRHTILSTI 102
+F W R+ L V G+ V +T + +E + K+IE +C+
Sbjct: 14 DFSLWKTRIMAHLSVIGLKDVVIGKTITPLTAEEEEDPEKKIE-------LCQ------- 59
Query: 103 SDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIKVQINEYHQ 162
A++ W+ ++ + + + ++ ++M+E+K+I I+++ +
Sbjct: 60 ------------TAQEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLK 107
Query: 163 LIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKH--KQMSLNDLI-----RHII 215
++ +L I + D A L+ L + +D + +K+ + +++ L+D++ +
Sbjct: 108 IVADLNHLQIEVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDDVMVGARDKERE 167
Query: 216 IEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPNFKANLGECYVCG 275
+ +R A+ N Q N K R +KS +V C++CG
Sbjct: 168 LSQNNRPVAEGHYARGRPEGKNNNQGNKGKN-RSRSKSADGKRV-----------CWICG 215
Query: 276 KKGH---KAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVN--DVQKW 330
K+GH + Y R + Q + + D ++ + LV D +W
Sbjct: 216 KEGHFKKQCYKWLERNKSKQQGSDVGESSLAKSSEIFDPAMVIMATGETLLVTGRDADEW 275
Query: 331 VVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSD 390
+D+G + H+ +R+ F + + V +G+ + V G G + ++ G + L+D
Sbjct: 276 FLDTGCSFHMTPRRDLFKDFKELSSGF--VKMGNDTYSPVKGIGSIKIRNNDGTQVILTD 333
Query: 391 VLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVL 442
V VP + NLIS+ L G + + + K + G + L++L
Sbjct: 334 VRYVPNMARNLISLGTLEDKGCWFKSQDGVLKIVKGCSTILKGQKRETLYIL 385
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 74.7 bits (182), Expect = 9e-14
Identities = 67/346 (19%), Positives = 151/346 (43%), Gaps = 27/346 (7%)
Query: 78 KSDAKQIEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKF 137
K DA ++ + +K ++ I ++D + K A + W+ ++ + + +
Sbjct: 34 KRDADEVARLERCDKA-KNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVY 92
Query: 138 VVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQ 197
++ ++M+E+K+I I+++ +++ +L I + D A L+ L + +D +
Sbjct: 93 TQLSFYTFKMQENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLVET 152
Query: 198 LKHKH--KQMSLNDLI-----RHIIIEDTSRKECGAARAKALESRANLIQNNARKQQRYE 250
+K+ + +++ L+D++ + + +R A+ N Q N K R
Sbjct: 153 MKYSNSREKLRLDDVMVAARDKERELSQNNRPVVEGHFARGRPDGKNNNQGNKGK-NRSR 211
Query: 251 NKSDHALKVTNPNFKANLGECYVCGKKGH---KAYHCRYRKVNGQPPKPKANLTQGDDKK 307
+KS +V C++CGK+GH + Y R + Q + +
Sbjct: 212 SKSADGKRV-----------CWICGKEGHFKKQCYKWIERNKSKQQGSDNGESSLAKSTE 260
Query: 308 YDDDDVIAAVISKVHLVND--VQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDS 365
+ ++ + +V D +WV+D+G + H+ +++ F + + V +G+
Sbjct: 261 AFNPAMVLLATDETLVVTDSIANEWVLDTGCSFHMTPRKDWFKDFKEL--SSGYVKMGND 318
Query: 366 RTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVG 411
+ V G G + ++ + G + L+DV +P + NLIS+ L G
Sbjct: 319 TYSPVKGIGSIKIRNSDGSQVILTDVRYMPNMTRNLISLGTLEDRG 364
>At4g10460 putative retrotransposon
Length = 1230
Score = 72.8 bits (177), Expect = 4e-13
Identities = 84/409 (20%), Positives = 170/409 (41%), Gaps = 60/409 (14%)
Query: 41 IEVFKGQ-NFRRWNERVFTLLDVHGVASVLTDVK----PDEAKSDAKQIEKWNNA----- 90
+E F G ++ W E++ +D+ G+ L + + P E++ + K+ EK +
Sbjct: 8 MEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALMEE 67
Query: 91 -NKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKE 149
+ R TI+ ++SD + K A + + ++ Y ++ + ++ ++M+E
Sbjct: 68 KRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQE 127
Query: 150 DKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLND 209
+ ++ I+E+ +LI +L+ N+++ D A L+ L +D K LK+ + +L+
Sbjct: 128 NLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS- 186
Query: 210 LIRHIIIEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPNFKANLG 269
+ ++ A +K LE +N + + Y ++ K N G
Sbjct: 187 -VDEVV---------AAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKGNKG 236
Query: 270 E----------CYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDK--KYDDDDVIAAV 317
C++CG++GH C + K +A+ ++G+ K + +
Sbjct: 237 RSRSRSKGWKGCWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSEGSGYY 296
Query: 318 ISKVHLVNDV---QKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKG 374
+S+ DV +WV+D+G H+ K+E F D V +G+ T+
Sbjct: 297 VSEALHSTDVNLGNEWVMDTGCNYHMTHKKEWFEELSE--DAGGTVRMGNKSTSKFR--- 351
Query: 375 KVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVM 423
V +P + NL+S+ L + G SFES V+
Sbjct: 352 ----------------VKYIPDMDRNLLSMGTLEEHG--YSFESKNGVL 382
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,990,372
Number of Sequences: 26719
Number of extensions: 424016
Number of successful extensions: 1553
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 1331
Number of HSP's gapped (non-prelim): 169
length of query: 443
length of database: 11,318,596
effective HSP length: 102
effective length of query: 341
effective length of database: 8,593,258
effective search space: 2930300978
effective search space used: 2930300978
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC139343.2


