

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138580.8 - phase: 0
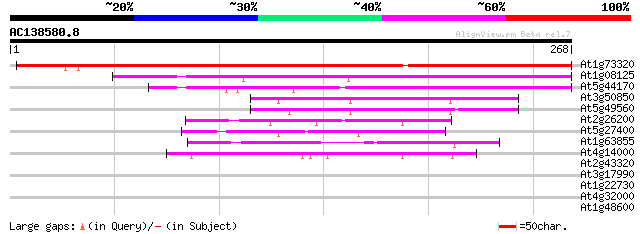
(268 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g73320 unknown protein 352 1e-97
At1g08125 DNA ligase 133 9e-32
At5g44170 unknown protein 74 6e-14
At3g50850 unknown protein 69 3e-12
At5g49560 putative protein 68 4e-12
At2g26200 unknown protein 62 2e-10
At5g27400 unknown protein 57 8e-09
At1g63855 putative protein 48 6e-06
At4g14000 unknown protein 45 3e-05
At2g43320 unknown protein 40 0.001
At3g17990 unknown protein 30 1.3
At1g22730 putative topoisomerase 28 3.9
At4g32000 serine/threonine protein kinase like protein 28 6.7
At1g48600 phosphoethanolamine N-methyltransferase, putative 27 8.7
>At1g73320 unknown protein
Length = 314
Score = 352 bits (903), Expect = 1e-97
Identities = 181/275 (65%), Positives = 211/275 (75%), Gaps = 12/275 (4%)
Query: 4 EEENATAAMVKLGSYGGSVMLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 53
+E+ A + ++ YGG V++V P ES AA E M++W IQ PT PN
Sbjct: 27 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 86
Query: 54 VAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQG 113
VAQSSL+LRLD+CGHSLSILQSP SL PGVTGSVMWDSG+VLGKFLEHSVDS +L L+G
Sbjct: 87 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 146
Query: 114 KKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTW 173
KKIVELGSGCGLVGCIAALLGG +LTDLPDR+RLL+KNI+TN+ + RGS EL W
Sbjct: 147 KKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAIVQELVW 206
Query: 174 GDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYF 233
GDDPD +LI+P PDY GSDV+YSE AV L++TL QL TTIFL+GELRNDA+LEYF
Sbjct: 207 GDDPDPDLIEPFPDY--GSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRNDAVLEYF 264
Query: 234 LEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
LE A+ DF IGRV+QT WHPDY S+RVVLYVL KK
Sbjct: 265 LETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 299
>At1g08125 DNA ligase
Length = 315
Score = 133 bits (335), Expect = 9e-32
Identities = 73/228 (32%), Positives = 125/228 (54%), Gaps = 13/228 (5%)
Query: 50 PNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGML 109
P+ + S+ + ++ GH L Q P+S G+ +WD+ +V K+L + G
Sbjct: 3 PDRLNSPSTCTVTIEVLGHELDFAQDPNSKH----LGTTVWDASMVFAKYLGKNSRKGRF 58
Query: 110 V---LQGKKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHI------ 160
L+GK+ +ELG+GCG+ G A+LG +V+ TD + + LL++N+E N I
Sbjct: 59 SSSKLKGKRAIELGAGCGVAGFALAMLGCDVVTTDQKEVLPLLKRNVEWNTSRIVQMNPG 118
Query: 161 SLRGSITATELTWGDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFL 220
S GS+ EL WG++ ++P DY++G+DVVYSE + LL T+ LSGP TT+ L
Sbjct: 119 SAFGSLRVAELDWGNEDHITAVEPPFDYVIGTDVVYSEQLLEPLLRTILALSGPKTTVML 178
Query: 221 AGELRNDAILEYFLEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
E+R+ + E L+ ++F + + ++ +Y + LY++ +K
Sbjct: 179 GYEIRSTVVHEKMLQMWKDNFEVKTIPRSKMDGEYQDPSIHLYIMAQK 226
>At5g44170 unknown protein
Length = 234
Score = 74.3 bits (181), Expect = 6e-14
Identities = 58/216 (26%), Positives = 99/216 (44%), Gaps = 20/216 (9%)
Query: 67 GHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEH--SVDSG-----------MLVLQG 113
G LSI Q S+ G+ +W ++L KF E ++DS + +
Sbjct: 15 GTKLSIQQDNGSMH----VGTSVWPCSLILSKFAERWSTLDSSSSTTSPNPYAELFDFRR 70
Query: 114 KKIVELGSGCGLVGCIAALLG-GEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELT 172
++ +ELG+GCG+ G LLG E++LTD+ M L+ N++ N +L S+ + +
Sbjct: 71 RRGIELGTGCGVAGMAFYLLGLTEIVLTDIAPVMPALKHNLKRNKT--ALGKSLKTSIVY 128
Query: 173 WGDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEY 232
W + + P D ++ +DVVY E +V L+ + L + + L ++R+ +
Sbjct: 129 WNNRDQISALKPPFDLVIAADVVYIEESVGQLVTAMELLVADDGAVLLGYQIRSPEADKL 188
Query: 233 FLEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
F E F I +V H DY +Y+ KK
Sbjct: 189 FWELCDIVFKIEKVPHEHLHSDYAYEETDVYIFRKK 224
>At3g50850 unknown protein
Length = 251
Score = 68.6 bits (166), Expect = 3e-12
Identities = 48/132 (36%), Positives = 66/132 (49%), Gaps = 4/132 (3%)
Query: 116 IVELGSGCGLVG-CIAALLGGEVILTDLPDRMRLLRKNIETNMKHIS-LRGSITATELTW 173
IVELGSG G+VG AA LG V +TDLP+ + L+ N + N + ++ G + L W
Sbjct: 92 IVELGSGTGIVGIAAAATLGANVTVTDLPNVIENLKFNADANAQVVAKFGGKVHVASLRW 151
Query: 174 GDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLG--QLSGPNTTIFLAGELRNDAILE 231
G+ D E + D IL SDVVY LL+TL L G + +FL L+
Sbjct: 152 GEIDDVESLGQNVDLILASDVVYHVHLYEPLLKTLRFLLLEGSSERVFLMAHLKRWKKES 211
Query: 232 YFLEAAMNDFTI 243
F + A F +
Sbjct: 212 IFFKKARRFFDV 223
>At5g49560 putative protein
Length = 274
Score = 68.2 bits (165), Expect = 4e-12
Identities = 51/132 (38%), Positives = 65/132 (48%), Gaps = 5/132 (3%)
Query: 116 IVELGSGCGLVGCIAAL-LGGEVILTDLPDRMRLLRKNIETNMKHIS-LRGSITATELTW 173
I+ELGSG GLVG AA+ L V +TDLP + L N E N + + G + L W
Sbjct: 109 ILELGSGTGLVGIAAAITLSANVTVTDLPHVLDNLNFNAEANAEIVERFGGKVNVAPLRW 168
Query: 174 GDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLG--QLSGPNTTIFLAGELRNDAILE 231
G+ D E++ D IL SDVVY + LL+TL QL G IFL LR
Sbjct: 169 GEADDVEVLGQNVDLILASDVVYHDHLYEPLLKTLRLMQLEG-KRLIFLMAHLRRWKKES 227
Query: 232 YFLEAAMNDFTI 243
F + A F +
Sbjct: 228 VFFKKARKLFDV 239
>At2g26200 unknown protein
Length = 565
Score = 62.4 bits (150), Expect = 2e-10
Identities = 41/133 (30%), Positives = 71/133 (52%), Gaps = 11/133 (8%)
Query: 85 TGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGC-GLVGCIAALLGGEVILTDLP 143
TG ++W+S ++ L+ + + ++ GK+++ELG GC G+ +AA V+ TD
Sbjct: 353 TGLMLWESARLMASVLDRNPN----IVSGKRVLELGCGCTGICSMVAARSANLVVATDAD 408
Query: 144 DR-MRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTP----DYILGSDVVYSE 198
+ + LL +NI N++ SL G + + L WG+ E I + I+G+DV Y
Sbjct: 409 TKALTLLTENITMNLQS-SLLGKLKTSVLEWGNKEHIESIKRLACEGFEVIMGTDVTYVA 467
Query: 199 GAVVDLLETLGQL 211
A++ L ET +L
Sbjct: 468 EAIIPLFETAKEL 480
>At5g27400 unknown protein
Length = 369
Score = 57.4 bits (137), Expect = 8e-09
Identities = 43/139 (30%), Positives = 61/139 (42%), Gaps = 18/139 (12%)
Query: 83 GVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVG-CIAALLGGEVILTD 141
G TG +W S + L +F V S + K E+GSG G+VG C+A + EVILTD
Sbjct: 145 GDTGCSIWPSSLFLSEF----VLSFPELFANKACFEVGSGVGMVGICLAHVKAKEVILTD 200
Query: 142 LPDRMRLLRKNIETNMKHISLRGS------------ITATELTWGDDPDQELIDPTPDYI 189
D + L + H++ + T L W + EL PD +
Sbjct: 201 -GDLLTLSNMKLNLERNHLNYDDEFLKQPGEAQSTRVKCTHLPWETASESELSQYRPDIV 259
Query: 190 LGSDVVYSEGAVVDLLETL 208
LG+DV+Y + LL L
Sbjct: 260 LGADVIYDPSCLPHLLRVL 278
>At1g63855 putative protein
Length = 205
Score = 47.8 bits (112), Expect = 6e-06
Identities = 42/151 (27%), Positives = 63/151 (40%), Gaps = 27/151 (17%)
Query: 86 GSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDLPDR 145
G +W ++L +++ + I+ELG+G L G +AA +G V LTD +
Sbjct: 34 GLFVWPCSVILAEYVWQHRSR----FRDSSILELGAGTSLPGLVAAKVGANVTLTDDATK 89
Query: 146 MRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTPDYILGSDVVYSEGAVVDLL 205
+L LTWG D ++D P+ ILG+DV+Y A DL
Sbjct: 90 PEVL--------------------GLTWG-VWDAPILDLRPNIILGADVLYDSSAFDDLF 128
Query: 206 ETLGQL--SGPNTTIFLAGELRNDAILEYFL 234
T+ L S P+ R+ L FL
Sbjct: 129 ATVSFLLQSSPDAVFITTYHNRSGHHLIEFL 159
>At4g14000 unknown protein
Length = 290
Score = 45.4 bits (106), Expect = 3e-05
Identities = 46/168 (27%), Positives = 75/168 (44%), Gaps = 20/168 (11%)
Query: 76 PSSLGKPGVT--GSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALL 133
P+S PGV G +W+ I L K LE +G L GK+++ELG G L G A L
Sbjct: 76 PNSDLVPGVYEGGLKLWEGSIDLVKALEKESQTGNLSFSGKRVLELGCGHALPGIYACLK 135
Query: 134 GGEVI-LTDL-PDRMRLLR-KNIETNMKHISLRGSITATELTWGDDPDQELIDPTP---- 186
G + + D + +R L N+ N+ S S++ TE+ + E+ P
Sbjct: 136 GSDAVHFQDFNAEVLRCLTIPNLNANLSEKSSSVSVSETEVRFFAGEWSEVHQVLPLVNS 195
Query: 187 ----------DYILGSDVVYSEGAVVDLLETLGQ-LSGPNTTIFLAGE 223
D IL ++ +YS A E + + L+ P+ +++A +
Sbjct: 196 DGETNKKGGYDIILMAETIYSISAQKSQYELIKRCLAYPDGAVYMAAK 243
>At2g43320 unknown protein
Length = 351
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/49 (34%), Positives = 28/49 (56%)
Query: 90 WDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVI 138
W+S +VL L++ + G L +GK+++ELG G+ G A L G +
Sbjct: 92 WESSVVLVNVLKNEIRDGQLSFRGKRVLELGCNFGVPGIFACLKGASSV 140
>At3g17990 unknown protein
Length = 165
Score = 30.0 bits (66), Expect = 1.3
Identities = 25/95 (26%), Positives = 45/95 (47%), Gaps = 8/95 (8%)
Query: 112 QGKKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATEL 171
+GK ++ELG+G G A GE+I D D + ++I + K++ ++
Sbjct: 26 EGKSVLELGAGIGRFTGELAQKAGELIALDFIDNVIKKNESINGHYKNV----KFMCADV 81
Query: 172 TWGDDPDQELIDPTPDYILGS-DVVYSEGAVVDLL 205
T PD ++ D + D I + ++Y V+LL
Sbjct: 82 T---SPDLKITDGSLDLIFSNWLLMYLSDKEVELL 113
>At1g22730 putative topoisomerase
Length = 693
Score = 28.5 bits (62), Expect = 3.9
Identities = 30/117 (25%), Positives = 49/117 (41%), Gaps = 20/117 (17%)
Query: 128 CIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTPD 187
CI+ L L LP + +++ + +H ++ + +E TWG + D +L DP D
Sbjct: 15 CISQLKISSSSLDPLPQAN--MAEDLTKSRRHSPIK--VEGSEETWGVEDDDDLTDPIFD 70
Query: 188 YILG---SDVV---------YSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEY 232
I G SD Y + A V + E G N + + EL+ + EY
Sbjct: 71 TIEGNGHSDPTSCFDADLSEYKKKATVIVEEYF----GTNDVVSVVNELKELGMAEY 123
>At4g32000 serine/threonine protein kinase like protein
Length = 379
Score = 27.7 bits (60), Expect = 6.7
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 2/63 (3%)
Query: 69 SLSILQSPSSLGK--PGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLV 126
S+S SP+S P T + GI +GK EH +D+ +L I G LV
Sbjct: 22 SVSAFTSPASQPSLSPVYTSMASFSPGIHMGKGQEHKLDAHKKLLIALIITSSSLGLILV 81
Query: 127 GCI 129
C+
Sbjct: 82 SCL 84
>At1g48600 phosphoethanolamine N-methyltransferase, putative
Length = 475
Score = 27.3 bits (59), Expect = 8.7
Identities = 17/61 (27%), Positives = 31/61 (49%), Gaps = 1/61 (1%)
Query: 112 QGKKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHIS-LRGSITATE 170
+GK ++ELG+G G A GEVI D + +++ + K+I + +T+ +
Sbjct: 37 EGKSVLELGAGIGRFTGELAQKAGEVIALDFIESAIQKNESVNGHYKNIKFMCADVTSPD 96
Query: 171 L 171
L
Sbjct: 97 L 97
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,991,109
Number of Sequences: 26719
Number of extensions: 248573
Number of successful extensions: 612
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 589
Number of HSP's gapped (non-prelim): 14
length of query: 268
length of database: 11,318,596
effective HSP length: 98
effective length of query: 170
effective length of database: 8,700,134
effective search space: 1479022780
effective search space used: 1479022780
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC138580.8


