

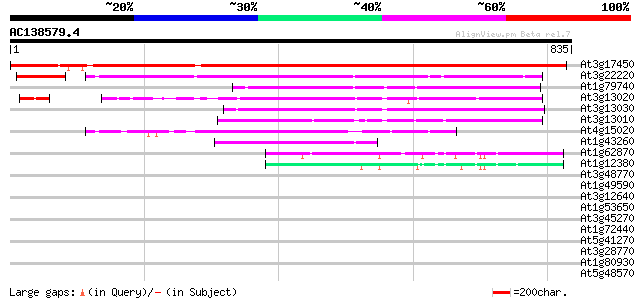
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138579.4 + phase: 0
(835 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g17450 unknown protein 948 0.0
At3g22220 hypothetical protein 342 5e-94
At1g79740 hypothetical protein 251 1e-66
At3g13020 hypothetical protein 246 5e-65
At3g13030 unknown protein (At3g13030) 211 1e-54
At3g13010 hypothetical protein 201 1e-51
At4g15020 putative protein 200 3e-51
At1g43260 hypothetical protein 144 2e-34
At1g62870 unknown protein 50 5e-06
At1g12380 hypothetical protein 44 3e-04
At3g48770 hypothetical protein 42 0.002
At1g49590 unknown protein 35 0.17
At3g12640 hypothetical protein 35 0.23
At1g53650 RNA-binding protein, putative 34 0.39
At3g45270 putative protein 33 0.66
At1g72440 hypothetical protein 33 0.66
At5g41270 unknown protein 32 1.9
At3g28770 hypothetical protein 32 1.9
At1g80930 unknown protein 31 2.5
At5g48570 peptidylprolyl isomerase 31 3.3
>At3g17450 unknown protein
Length = 877
Score = 948 bits (2451), Expect = 0.0
Identities = 476/845 (56%), Positives = 627/845 (73%), Gaps = 32/845 (3%)
Query: 1 MAPIRSTGFVDPGWDHGIAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEKA 60
MAP S G VDPGW+HG+AQD+RKKKV+CNYCGK+VSGGIYRLKQHLARVSGEVTYC+K+
Sbjct: 1 MAPPGSIGVVDPGWEHGVAQDQRKKKVKCNYCGKIVSGGIYRLKQHLARVSGEVTYCDKS 60
Query: 61 PEEVYLKMKENLEGCRSNKKQKQVD---AQAYMNFQ--SNDDEDDEEQVGC---RSKGKQ 112
PEEV ++MKENL RS KK +Q + Q+ +F +NDDE DEE+ C RSKGK
Sbjct: 61 PEEVCMRMKENL--VRSTKKLRQSEDNSGQSCSSFHQSNNDDEADEEERRCWSIRSKGKL 118
Query: 113 -LMDGRNVSVNLTPLRSLGYVDPGWEHGVAQDERKKKVKCSYCEKVVSGGINRFKQHLAR 171
L DG + LRS GY+DPGWEHG+AQDERKKKVKC+YC K+VSGGINRFKQHLAR
Sbjct: 119 GLSDG-------SLLRSSGYIDPGWEHGIAQDERKKKVKCNYCNKIVSGGINRFKQHLAR 171
Query: 172 IPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAK-DLMPFYPKSDNEDDEYEQQED 230
IPGEVAPCK+APEEVY+KIKENMKWHR GKR +P+ + + F S + D E ++++
Sbjct: 172 IPGEVAPCKTAPEEVYVKIKENMKWHRAGKRQNRPDDEMGALTFRTVSQDPDQEEDREDH 231
Query: 231 TLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCK 290
+ +++ L+ + R+SKD K+F + + E +R+R+ F Q+ + K
Sbjct: 232 DFYPTSQDRLMLGNGRFSKDKRKSFDSTNMRSVSEAKTKRARMIPF-------QSPSSSK 284
Query: 291 QLKVKTGPTKKL--RKEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSS 348
Q K+ + + ++ RK+V SSI KF H G+P +AA+S+YF KM+EL G YG+G PSS
Sbjct: 285 QRKLYSSCSNRVVSRKDVTSSISKFLHHVGVPTEAANSLYFQKMIELIGMYGEGFVVPSS 344
Query: 349 QLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFV 408
QL SGR LQEE+++IK+YL EY++SW +TGCSIMAD+W + +G+ +I+FLVS P GVYF
Sbjct: 345 QLFSGRLLQEEMSTIKSYLREYRSSWVVTGCSIMADTWTNTEGKKMISFLVSCPRGVYFH 404
Query: 409 SSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTP 468
SS+DAT++VEDA LFK LDK+V+++GEENVVQVIT+NT +++AGK+LEE+R+NL+WTP
Sbjct: 405 SSIDATDIVEDALSLFKCLDKLVDDIGEENVVQVITQNTAIFRSAGKLLEEKRKNLYWTP 464
Query: 469 CAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQ 528
CAI+C VLEDF K+ V EC+EK Q+IT+ IYNQ WLLNLMK+EFT G +LL+PA +
Sbjct: 465 CAIHCTELVLEDFSKLEFVSECLEKAQRITRFIYNQTWLLNLMKNEFTQGLDLLRPAVMR 524
Query: 529 CASSFATLQNLLDHRVSLRRMFLSNKW-MSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRN 587
AS F TLQ+L+DH+ SLR +F S+ W +S + S +G+EV+K+VL+ FWKK+Q V
Sbjct: 525 HASGFTTLQSLMDHKASLRGLFQSDGWILSQTAAKSEEGREVEKMVLSAVFWKKVQYVLK 584
Query: 588 SVYPILQVFQKVS-SGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSL 646
SV P++QV ++ G+ LSMPY Y + AK+AIKSIH DDARKY PFW+VI+ N L
Sbjct: 585 SVDPVMQVIHMINDGGDRLSMPYAYGYMCCAKMAIKSIHSDDARKYGPFWRVIEYRWNPL 644
Query: 647 FCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQ 706
F HPLY+AAYF NP+Y+YR DF++ S+VVRG+NECIVRLE DN RRI+A MQIP Y A+
Sbjct: 645 FHHPLYVAAYFFNPAYKYRPDFMAQSEVVRGVNECIVRLEPDNTRRITALMQIPDYTCAK 704
Query: 707 DDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSFACEHDGSMYDQIYS 766
DFGT++AI TRT L+P+AWWQQHGISCLELQR+AVRILS TCSS CE S+YDQ+ S
Sbjct: 705 ADFGTDIAIGTRTELDPSAWWQQHGISCLELQRVAVRILSHTCSSVGCEPKWSVYDQVNS 764
Query: 767 KRKNRLSQKKLNDIMYVHYNLRLRECQVRKR--SRESKSTSAENVLQEHLLGDWIVDTTA 824
+ +++ +K D+ YVHYNLRLRE Q+++R + + + L + LL DW+V +
Sbjct: 765 QCQSQFGKKSTKDLTYVHYNLRLREKQLKQRLHYEDEPPPTLNHALLDRLLPDWLVTSEK 824
Query: 825 QSSDS 829
+ ++
Sbjct: 825 EEEEA 829
>At3g22220 hypothetical protein
Length = 759
Score = 342 bits (877), Expect = 5e-94
Identities = 216/686 (31%), Positives = 358/686 (51%), Gaps = 20/686 (2%)
Query: 114 MDGRNVSVNLTPLRSLGYVDPGWEH-GVAQDERKKKVKCSYCEKVVSGG-INRFKQHLAR 171
MD V LTP + D W+H V + + +++C YC K+ GG I R K+HLA
Sbjct: 1 MDSDLEPVALTPQKQ----DSAWKHCEVYKYGDRVQMRCLYCRKMFKGGGITRVKEHLAG 56
Query: 172 IPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDLMP--FYPKSDNEDDEYEQQE 229
G+ C P+EV L +++ + +R R+ + + +P ++P + E +
Sbjct: 57 KKGQGTICDQVPDEVRLFLQQCIDGTVRRQRKRRKSSPEPLPIAYFPPCEVETQVAASSD 116
Query: 230 DTLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTC 289
+ + + + + + +T++ +N L +D + N
Sbjct: 117 VNNGFKSPSSDVVVGQSTGRTKQRTYRSRKNNAFERNDLANVEVD----RDMDNLIPVAI 172
Query: 290 KQLKVKTGPTKKLR-KEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSS 348
+K PT K R K V ++ +F G AA+SV ++ G G++ P+
Sbjct: 173 SSVKNIVHPTSKEREKTVHMAMGRFLFDIGADFDAANSVNVQPFIDAIVSGGFGVSIPTH 232
Query: 349 QLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFV 408
+ + G L+ + +K + E K W TGCS++ +G I+ FLV P V F+
Sbjct: 233 EDLRGWILKSCVEEVKKEIDECKTLWKRTGCSVLVQELNSNEGPLILKFLVYCPEKVVFL 292
Query: 409 SSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTP 468
SVDA+ +++ L++LL +VVEE+G+ NVVQVIT+ +Y AAGK L + +L+W P
Sbjct: 293 KSVDASEILDSEDKLYELLKEVVEEIGDTNVVQVITKCEDHYAAAGKKLMDVYPSLYWVP 352
Query: 469 CAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQ 528
CA +CI+++LE+F K+ + E +E+ + +T++IYN +LNLM+ +FT GN++++P T
Sbjct: 353 CAAHCIDKMLEEFGKMDWIREIIEQARTVTRIIYNHSGVLNLMR-KFTFGNDIVQPVCTS 411
Query: 529 CASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNS 588
A++F T+ + D + L+ M S++W +S + G + + + + FWK + +
Sbjct: 412 SATNFTTMGRIADLKPYLQAMVTSSEWNDCSYSKEAGGLAMTETINDEDFWKALTLANHI 471
Query: 589 VYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFC 648
PIL+V + V S +M Y+Y +YRAK AIK+ + +Y +WK+IDR
Sbjct: 472 TAPILRVLRIVCSERKPAMGYVYAAMYRAKEAIKT-NLAHREEYIVYWKIIDRW---WLQ 527
Query: 649 HPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDD 708
PLY A ++LNP + Y D S++ + +CI +L D + I Y +A
Sbjct: 528 QPLYAAGFYLNPKFFYSIDEEMRSEIHLAVVDCIEKLVPDVNIQDIVIKDINSYKNAVGI 587
Query: 709 FGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTC-SSFACEHDGSMYDQIYSK 767
FG LAI R + PA WW +G SCL L R A+RILSQTC SS + + QIY +
Sbjct: 588 FGRNLAIRARDTMLPAEWWSTYGESCLNLSRFAIRILSQTCSSSIGSVRNLTSISQIY-E 646
Query: 768 RKNRLSQKKLNDIMYVHYNLRLRECQ 793
KN + +++LND+++V YN+RLR +
Sbjct: 647 SKNSIERQRLNDLVFVQYNMRLRRIE 672
Score = 49.7 bits (117), Expect = 7e-06
Identities = 27/76 (35%), Positives = 46/76 (60%), Gaps = 4/76 (5%)
Query: 11 DPGWDH-GIAQDERKKKVRCNYCGKVVSGG-IYRLKQHLARVSGEVTYCEKAPEEVYLKM 68
D W H + + + ++RC YC K+ GG I R+K+HLA G+ T C++ P+EV L +
Sbjct: 16 DSAWKHCEVYKYGDRVQMRCLYCRKMFKGGGITRVKEHLAGKKGQGTICDQVPDEVRLFL 75
Query: 69 KENLEGC--RSNKKQK 82
++ ++G R K++K
Sbjct: 76 QQCIDGTVRRQRKRRK 91
>At1g79740 hypothetical protein
Length = 518
Score = 251 bits (641), Expect = 1e-66
Identities = 140/458 (30%), Positives = 258/458 (55%), Gaps = 7/458 (1%)
Query: 332 MLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQG 391
ML+ + G G PS + +L + I L + + W TGC+I+A++W D +
Sbjct: 1 MLDAVAKCGPGFVAPSPKT---EWLDRVKSDISLQLKDTEKEWVTTGCTIIAEAWTDNKS 57
Query: 392 RTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYK 451
R +INF VSSP ++F SVDA++ +++ L L D V++++G+E++VQ+I +N+ Y
Sbjct: 58 RALINFSVSSPSRIFFHKSVDASSYFKNSKCLADLFDSVIQDIGQEHIVQIIMDNSFCYT 117
Query: 452 AAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLM 511
L + +F +PCA C+N +LE+F K+ V +C+ + Q I+K +YN +L+L+
Sbjct: 118 GISNHLLQNYATIFVSPCASQCLNIILEEFSKVDWVNQCISQAQVISKFVYNNSPVLDLL 177
Query: 512 KSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQK 571
+ + T G ++++ T+ S+F +LQ+++ + L+ MF ++ ++ ++ Q
Sbjct: 178 R-KLTGGQDIIRSGVTRSVSNFLSLQSMMKQKARLKHMFNCPEYTTN--TNKPQSISCVN 234
Query: 572 IVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARK 631
I+ + FW+ ++ PIL+V ++VS+G+ ++ IY + +AK +I++ + D K
Sbjct: 235 ILEDNDFWRAVEESVAISEPILKVLREVSTGKP-AVGSIYELMSKAKESIRTYYIMDENK 293
Query: 632 YEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMR 691
++ F ++D + PL+ AA FLNPS +Y + + + + + +L +
Sbjct: 294 HKVFSDIVDTNWCEHLHSPLHAAAAFLNPSIQYNPEIKFLTSLKEDFFKVLEKLLPTSDL 353
Query: 692 RISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSS 751
R + QI + A+ FG LA+ R + P WW+Q G S LQR+A+RILSQ CS
Sbjct: 354 RRDITNQIFTFTRAKGMFGCNLAMEARDSVSPGLWWEQFGDSAPVLQRVAIRILSQVCSG 413
Query: 752 FACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRL 789
+ E S + Q++ +R+N++ ++ LN + YV+ NL+L
Sbjct: 414 YNLERQWSTFQQMHWERRNKIDREILNKLAYVNQNLKL 451
>At3g13020 hypothetical protein
Length = 605
Score = 246 bits (627), Expect = 5e-65
Identities = 178/666 (26%), Positives = 308/666 (45%), Gaps = 80/666 (12%)
Query: 137 EHGVAQDERKKKVKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSAPEEVYLKIKENMKW 196
EHG+ D++K +VKC+YC K ++ +R K HL + +V C ++V L ++E +
Sbjct: 10 EHGICVDKKKSRVKCNYCGKEMNS-FHRLKHHLGAVGTDVTHC----DQVSLTLRETFRT 64
Query: 197 HRTGKR--HRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKT 254
+ + P+ K + F Q D S+ KT
Sbjct: 65 MLMEDKSGYTTPKTKRVGKF------------QMAD-----------------SRKRRKT 95
Query: 255 FKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCKQLKVKTGPTKKLRKEVFSSICKFF 314
S + SPE +D NQ+L + K K I +FF
Sbjct: 96 EDSSSKSVSPEQGNVAVEVD--------NQDLLSSKAQKC---------------IGRFF 132
Query: 315 CHAGIPLQAADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASW 374
+ L A DS F +M+ G G P S ++GR LQE + +++Y+ K SW
Sbjct: 133 YEHCVDLSAVDSPCFKEMMMALGV---GQKIPDSHDLNGRLLQEAMKEVQDYVKNIKDSW 189
Query: 375 AITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEEL 434
ITGCSI+ D+W D +G +++F+ P G ++ S+D + V D T L L++ +VEE+
Sbjct: 190 KITGCSILLDAWIDPKGHDLVSFVADCPAGPVYLKSIDVSVVKNDVTALLSLVNGLVEEV 249
Query: 435 GEENVVQVITENTPNYKA-AGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEK 493
G NV Q+I +T + GK+ R +FW+ +C +L K+R + ++K
Sbjct: 250 GVHNVTQIIACSTSGWVGELGKLFSGHDREVFWSVSLSHCFELMLVKIGKMRSFGDILDK 309
Query: 494 GQKITKLIYNQIWLLNLMKSEFTHGNEL-LKPAGTQCASSFATLQNLLDHRVSLRRMFLS 552
I + I N L + + + +HG ++ + + + + L+++ + +L MF S
Sbjct: 310 VNTIWEFINNNPSALKIYRDQ-SHGKDITVSSSEFEFVKPYLILKSVFKAKKNLAAMFAS 368
Query: 553 NKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPI---LQVFQKVSSGESLSMPY 609
+ W +GK V +V + +FW+ ++ + P+ L++F + + + Y
Sbjct: 369 SVW------KKEEGKSVSNLVNDSSFWEAVEEILKCTSPLTDGLRLFSNADNNQHVG--Y 420
Query: 610 IYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFV 669
IY+ L KL+IK D+ + Y W VID N +PL+ A Y+LNP+ Y DF
Sbjct: 421 IYDTLDGIKLSIKKEFNDEKKHYLTLWDVIDDVWNKHLHNPLHAAGYYLNPTSFYSTDFH 480
Query: 670 SHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQ 729
+V GL +V + + +I++ Q+ Y +D F +G+ P WW +
Sbjct: 481 LDPEVSSGLTHSLVHVAKEGQIKIAS--QLDRYRLGKDCFNEASQPDQISGISPIDWWTE 538
Query: 730 HGISCLELQRIAVRILSQTCSSFA-CEHDGSMYDQ-IYSKRKNRLSQKKLNDIMYVHYNL 787
ELQ A++ILSQTC + + S+ ++ + ++ + +K L ++ +VHYNL
Sbjct: 539 KASQHPELQSFAIKILSQTCEGASRYKLKRSLAEKLLLTEGMSHCERKHLEELAFVHYNL 598
Query: 788 RLRECQ 793
L+ C+
Sbjct: 599 HLQSCK 604
Score = 52.4 bits (124), Expect = 1e-06
Identities = 21/45 (46%), Positives = 33/45 (72%), Gaps = 1/45 (2%)
Query: 15 DHGIAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEK 59
+HGI D++K +V+CNYCGK ++ +RLK HL V +VT+C++
Sbjct: 10 EHGICVDKKKSRVKCNYCGKEMN-SFHRLKHHLGAVGTDVTHCDQ 53
>At3g13030 unknown protein (At3g13030)
Length = 544
Score = 211 bits (538), Expect = 1e-54
Identities = 133/482 (27%), Positives = 243/482 (49%), Gaps = 14/482 (2%)
Query: 319 IPLQAADSVYFHKMLELAGQYGQ-GLACPSSQLISGRFLQEEINSIKNYLAEYKASWAIT 377
+ L A D+ F +M+ + G GQ GL ++G LQ+ + +++ + + K SWAIT
Sbjct: 62 VNLSAVDAPCFKEMMTVDG--GQMGLESSDCHDLNGWRLQDALEEVQDRVEKIKESWAIT 119
Query: 378 GCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEE 437
GCSI+ D+W D +GR ++ F+ P G+ ++ S D ++ +D T L L++ +VEE+G
Sbjct: 120 GCSILLDAWVDQKGRDLVTFVADCPAGLVYLISFDVSDFKDDVTALLSLVNGLVEEVGVR 179
Query: 438 NVVQVITENTPNYKAA-GKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQK 496
NV Q+I +T + G++ R +FW+ +C +L KIR + +K
Sbjct: 180 NVTQIIACSTSGWVGELGELFAGHDREVFWSVSVSHCFELMLVKISKIRSFGDIFDKVNN 239
Query: 497 ITKLIYNQIWLLNLMKSEFTHGNEL-LKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKW 555
I I N +LN+ + + HG ++ + + + + + L+++ + +L MF S+ W
Sbjct: 240 IWLFINNNPSVLNIFRDQ-CHGIDITVSSSEFEFVTPYLILESIFKAKKNLTAMFASSNW 298
Query: 556 MSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLY 615
++ Q + +V + +FW+ ++SV P++ S+ + + Y+Y+ +
Sbjct: 299 ------NNEQCIAISNLVSDSSFWETVESVLKCTSPLIHGLLLFSTANNQHLGYVYDTMD 352
Query: 616 RAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVV 675
K +I + Y+P W VID N +PL+ A YFLNP+ Y +F +VV
Sbjct: 353 SIKESIAREFNHKPQFYKPLWDVIDDVWNKHLHNPLHAAGYFLNPTAFYSTNFHLDIEVV 412
Query: 676 RGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCL 735
GL ++ + D + S QI Y +D F TG+ PA WW
Sbjct: 413 TGLISSLIHMVEDCHVQFKISTQIDMYRLGKDCFNEASQADQITGISPAEWWAHKASQYP 472
Query: 736 ELQRIAVRILSQTCSSFA-CEHDGSMYDQ-IYSKRKNRLSQKKLNDIMYVHYNLRLRECQ 793
ELQ +A++ILSQTC + + S+ ++ + S+ + ++ L+++++V YNL L+ +
Sbjct: 473 ELQSLAIKILSQTCEGASKYKLKRSLAEKLLLSEGMSNRERQHLDELVFVQYNLHLQSYK 532
Query: 794 VR 795
+
Sbjct: 533 AK 534
>At3g13010 hypothetical protein
Length = 572
Score = 201 bits (511), Expect = 1e-51
Identities = 140/492 (28%), Positives = 233/492 (46%), Gaps = 27/492 (5%)
Query: 310 ICKFFCHAGIPLQAADSVYFHKMLEL--AGQYGQGLACPSSQLISGRFLQEEINSIKNYL 367
+ +FF G+ A DS F KM+ + G G G P S+ ++G QE + +++ +
Sbjct: 99 VAQFFYEHGVDFSAVDSTSFKKMMMIKTVGGEGGGQMIPDSRDLNGWMFQEALKKVQDRV 158
Query: 368 AEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLL 427
E KASW ITGCSI+ D+W +GR ++ F+ P G ++ S D +++ D T L L+
Sbjct: 159 KEIKASWEITGCSILFDAWIGPKGRDLVTFVADCPAGAVYLKSADVSDIKTDVTALTSLV 218
Query: 428 DKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCV 487
+ +VEE+G NV Q+I +T + G + ++ +FW+ YC+ +L + K+
Sbjct: 219 NGIVEEVGVRNVTQIIACSTSGW--VGDLGKQLAGQVFWSVSLSYCLKLMLVEIGKMYSF 276
Query: 488 EECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLR 547
E+ EK + + LI N L + + N ++C F L+H +R
Sbjct: 277 EDIFEKVKLLLDLINNNPSFLYVFRE-----NSHKVDVSSEC--EFVMPYLTLEHIYWVR 329
Query: 548 R--MFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESL 605
R +F S +W QG + V + TFW+ + + S ++ + S G S
Sbjct: 330 RAGLFASPEW------KKEQGIAISSFVNDSTFWESLDKIVGSTSSLVHGWLWFSRG-SK 382
Query: 606 SMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDR--HCNSLFCHPLYLAAYFLNPSYR 663
+ Y Y+ + K + + + YEP W VID H N +PL+ A YFLNP
Sbjct: 383 HVAYAYHFIESIKKNVAWTFKYERQFYEPTWNVIDDVWHNNH---NPLHAAGYFLNPMAY 439
Query: 664 YRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEP 723
Y DF ++ V GL +V L + ++ Q+ Y + F G+ P
Sbjct: 440 YSDDFHTYQHVYTGLAFSLVHLVKEPHLQVKIGTQLDVYRYGRGCFMKASQAGQLNGVSP 499
Query: 724 AAWWQQHGISCLELQRIAVRILSQTCSSFA-CEHDGSMYDQ-IYSKRKNRLSQKKLNDIM 781
WW Q ELQ +AV+ILSQT + + S+ ++ + ++ + +K L ++
Sbjct: 500 VNWWTQKANQYPELQNLAVKILSQTSEGASRYKLKRSVAEKLLLTEGMSHCERKHLEELA 559
Query: 782 YVHYNLRLRECQ 793
VHYNL+L+ C+
Sbjct: 560 VVHYNLQLQSCK 571
Score = 33.9 bits (76), Expect = 0.39
Identities = 13/36 (36%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query: 148 KVKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSAP 183
+V+C++C+K ++ NR K HL + +V PC P
Sbjct: 20 RVQCTFCDKRMND-FNRLKYHLGNVCKDVTPCSQVP 54
Score = 32.0 bits (71), Expect = 1.5
Identities = 17/46 (36%), Positives = 25/46 (53%), Gaps = 4/46 (8%)
Query: 26 KVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEKAP---EEVYLKM 68
+V+C +C K ++ RLK HL V +VT C + P E + KM
Sbjct: 20 RVQCTFCDKRMND-FNRLKYHLGNVCKDVTPCSQVPLPIREAFFKM 64
>At4g15020 putative protein
Length = 522
Score = 200 bits (508), Expect = 3e-51
Identities = 160/565 (28%), Positives = 251/565 (44%), Gaps = 64/565 (11%)
Query: 114 MDGRNVSVNLTPLRSLGYVDPGWEH-GVAQDERKKKVKCSYCEKVVSGG-INRFKQHLAR 171
MD V LTP + D W+H + + + +++C YC K+ GG I R K
Sbjct: 1 MDAELEPVALTPQKQ----DNAWKHCEIYKYGDRLQMRCLYCRKMFKGGGITRVK----- 51
Query: 172 IPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQ-----PEAKDLMPFYP------KSDN 220
++ K + + ++ K H++ P D+M P KS
Sbjct: 52 ---DILLVKRDKQCIDGTVRRQRKRHKSSSEPLSVASLPPIEGDMMVVQPDVNDGFKSPG 108
Query: 221 EDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKH 280
D Q E L K+ R Y G +SN +
Sbjct: 109 SSDVVVQNESLLSGRTKQ------RTYRSKKNAFENGSASNNV-----------DLIGRD 151
Query: 281 PTNQNLQTCKQLKVKTGPTKKLRKE-VFSSICKFFCHAGIPLQAADSVYFHKMLELAGQY 339
N +K P+ + R+ + +I +F G A +SV F M++
Sbjct: 152 MDNLIPVAISSVKNIVHPSFRDRENTIHMAIGRFLFGIGADFDAVNSVNFQPMIDAIASG 211
Query: 340 GQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLV 399
G G++ P+ + G L+ + + + E KA W TGCSI+ + +G ++NFLV
Sbjct: 212 GFGVSAPTHDDLRGWILKNCVEEMAKEIDECKAMWKRTGCSILVEELNSDKGFKVLNFLV 271
Query: 400 SSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEE 459
P V F+ SVDA+ V+ A LF+LL ++VEE+G NVVQVIT+ Y AGK L
Sbjct: 272 YCPEKVVFLKSVDASEVLSSADKLFELLSELVEEVGSTNVVQVITKCDDYYVDAGKRLML 331
Query: 460 RRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGN 519
+L+W PCA +CI+Q+LE+F K+ + E +E+ Q IT+ +YN
Sbjct: 332 VYPSLYWVPCAAHCIDQMLEEFGKLGWISETIEQAQAITRFVYNH--------------- 376
Query: 520 ELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFW 579
A + A++FATL + + + +L+ M S +W +S G V + + FW
Sbjct: 377 ----SAFSSSATNFATLGRIAELKSNLQAMVTSAEWNECSYSEEPSGL-VMNALTDEAFW 431
Query: 580 KKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVI 639
K + V + P+L+ + V S + +M Y+Y LYRAK AIK+ H + Y +WK+I
Sbjct: 432 KAVALVNHLTSPLLRALRIVCSEKRPAMGYVYAALYRAKDAIKT-HLVNREDYIIYWKII 490
Query: 640 DRHCNSLFCHPLYLAAYFLNPSYRY 664
D ++ L +FLNP Y
Sbjct: 491 DHGGSNNSIFLCLLLGFFLNPKLFY 515
>At1g43260 hypothetical protein
Length = 294
Score = 144 bits (363), Expect = 2e-34
Identities = 78/243 (32%), Positives = 135/243 (55%), Gaps = 2/243 (0%)
Query: 306 VFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKN 365
V + ++ GIP A + +MLE+AGQ+G G+ PS + L+EE+ +K
Sbjct: 35 VHQYVARWVYSHGIPFNAIANDDLRRMLEVAGQFGPGVTPPSQYQLREPLLKEEVVRMKG 94
Query: 366 YLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFK 425
+ E + W + GCS+ DSW D + R+I+N ++ G F+SS D + Y+F
Sbjct: 95 LMEEQEDEWRVNGCSVTTDSWSDRKRRSIMNLCINCKEGTMFLSSKDCFDDSHTGEYIFA 154
Query: 426 LLDK-VVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKI 484
+++ ++ LG ++VVQV+T N N A K+L+E R +FWT CA + IN ++E K+
Sbjct: 155 YVNEYCIKNLGGDHVVQVVTNNATNNITAAKLLKEVRPTIFWTFCATHTINLMVEGISKL 214
Query: 485 RCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRV 544
+E ++ + T IY L++M+S FT ++++P AS+F TL++L++
Sbjct: 215 AMSDEIVKMAKAFTIFIYAHHQTLSMMRS-FTKRRDIVRPGIIGFASAFRTLKSLVEKEE 273
Query: 545 SLR 547
+L+
Sbjct: 274 NLK 276
>At1g62870 unknown protein
Length = 796
Score = 50.1 bits (118), Expect = 5e-06
Identities = 87/482 (18%), Positives = 196/482 (40%), Gaps = 51/482 (10%)
Query: 381 IMADSWR-DAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEEL---GE 436
I +D W+ D+ G ++N +V+ P+G N + Y ++L + V +
Sbjct: 293 IASDGWKFDSSGENLVNLIVNLPNGTSLYRRAVFVNGAVPSNYAEEVLWETVRGICGNSP 352
Query: 437 ENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLK-IRCVEECMEKGQ 495
+ V ++++ A + LE + + + C N ++ DF+K + + +
Sbjct: 353 QRCVGIVSDRF--MSKALRNLESQHQWMVNLSCQFQGFNSLIRDFVKELPLFKSVSQSCS 410
Query: 496 KITKLIYNQIWLLN-LMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRR---MFL 551
++ + + + N + K + E +S F L NLL+ +S R + +
Sbjct: 411 RLVNFVNSTAQIRNAVCKYQLQEQGETRMLHLPLDSSLFEPLYNLLEDVLSFARAIQLVM 470
Query: 552 SNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIY 611
+ + +EV ++V +V FW +++ +VY +L++ ++++ P +
Sbjct: 471 HDDVCKAVLMEDHMAREVGEMVGDVGFWNEVE----AVYLLLKLVKEMARRIEEERPLVG 526
Query: 612 N-----DLYRAKLA--IKSIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSY-- 662
D R+K+ + + R+ E K+++R + HP + AA+ L+P Y
Sbjct: 527 QCLPLWDELRSKIKDWYAKFNVVEERQVE---KIVERRFKKSY-HPAWAAAFILDPLYLI 582
Query: 663 -----RYRQDFVSHS-DVVRGLNECIVRLELDNMRRISASMQI---------PHYNSA-- 705
+Y F S + + +++ I RL + I A M++ P Y A
Sbjct: 583 KDSSGKYLPPFKCLSPEQEKDVDKLITRLVSRDEAHI-AMMELMKWRTEGLDPVYARAVQ 641
Query: 706 ---QDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSFACEHDGSMYD 762
+D ++ I+ W+ + L R+AVR++ +S + + S+
Sbjct: 642 MKERDPVSGKMRIANPQSSRLV--WETYLSEFRSLGRVAVRLIFLHATSCGFKCNSSVLR 699
Query: 763 QIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRSRESKSTSAENVLQEHLLGDWIVDT 822
+ S ++R + + ++++ N + A ++ +L D ++DT
Sbjct: 700 WVNSNGRSRAAVDRAQKLIFISANSKFERRDFSNEEERDAELLAMANGEDDVLNDVLIDT 759
Query: 823 TA 824
++
Sbjct: 760 SS 761
>At1g12380 hypothetical protein
Length = 793
Score = 44.3 bits (103), Expect = 3e-04
Identities = 88/497 (17%), Positives = 200/497 (39%), Gaps = 63/497 (12%)
Query: 381 IMADSWRDAQ-GRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENV 439
I +D W+ + G +++N +V+ P+G N + Y ++L + V+ + +
Sbjct: 306 ISSDGWKPGESGESLVNLIVNLPNGTSLYRRAVLVNGAVPSNYAEEVLLETVKGICGNSP 365
Query: 440 VQVITENTPNYKA-AGKMLEERRRNLFWTPCAIYCINQVLEDFLK-IRCVEECMEKGQKI 497
+ + + +K A + LE + + + C +N +++DF+K + + + ++
Sbjct: 366 QRCVGIVSDKFKTKALRNLESQHQWMVNLSCQFQGLNSLIKDFVKELPLFKSVSQNCVRL 425
Query: 498 TKLIYNQIWLLNL-MKSEFT-HGNELL------------------KPAGTQCASSFATLQ 537
K I N + N K + HG ++ +G+ + L
Sbjct: 426 AKFINNTAQIRNAHCKYQLQEHGESIMLRLPLHCYYDDERRSCSSSSSGSNKVCFYEPLF 485
Query: 538 NLLDHRVSLRR---MFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQ 594
NLL+ +S R + + + +EV+++V + FW ++++V + + +
Sbjct: 486 NLLEDVLSSARAIQLVVHDDACKVVLMEDHMAREVREMVGDEGFWNEVEAVHALIKLVKE 545
Query: 595 VFQKVSSGESL---SMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHPL 651
+ +++ + L +P ++++L RAK+ + + KV++R + HP
Sbjct: 546 MARRIEEEKLLVGQCLP-LWDEL-RAKVKDWDSKFNVGEGHVE--KVVERRFKKSY-HPA 600
Query: 652 YLAAYFLNPSYRYRQDFVSH--------SDVVRGLNECIVRLELDNMRRISASMQI---- 699
+ AA+ L+P Y R + + + +++ I RL + I A M++
Sbjct: 601 WAAAFILDPLYLIRDSSGKYLPPFKCLSPEQEKDVDKLITRLVSRDEAHI-ALMELMKWR 659
Query: 700 -----PHYNSA-----QDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRI--LSQ 747
P Y A +D ++ I+ W+ + L ++AVR+ L
Sbjct: 660 TEGLDPMYARAVQMKERDPVSGKMRIANPQSSRLV--WETYLSEFRSLGKVAVRLIFLHA 717
Query: 748 TCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRSRESKSTSAE 807
T F C + S+ + S ++ + + ++++ N + A
Sbjct: 718 TTGGFKC--NSSLLKWVNSNGRSHAAVDRAQKLIFISANSKFERRDFSNEEDRDAELLAM 775
Query: 808 NVLQEHLLGDWIVDTTA 824
+H+L D +VDT++
Sbjct: 776 ANGDDHMLNDVLVDTSS 792
>At3g48770 hypothetical protein
Length = 1899
Score = 41.6 bits (96), Expect = 0.002
Identities = 33/108 (30%), Positives = 51/108 (46%), Gaps = 10/108 (9%)
Query: 23 RKKKVRCNYCGK-VVSGGIYRLKQHLARVSGEVTYCEKAPEEVYLKMKENLEGCRSNKKQ 81
R+K + C +CGK V I R+K HLA ++ + C+K EV M+++LE + +K
Sbjct: 41 RRKVITCGFCGKQYVGSEINRMKHHLAGLNNR-SACQKVSSEVQHSMRKSLE--ENKEKP 97
Query: 82 KQVDAQAYMNFQSNDDEDDEEQVGCRSKGKQLMDGRNVSVNLTPLRSL 129
K++ D +DDE VG S R V + +SL
Sbjct: 98 KKIIV------VDLDMQDDEVNVGQSSHQSCSSSSRKRKVQHSMRKSL 139
Score = 32.0 bits (71), Expect = 1.5
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 2/52 (3%)
Query: 145 RKKKVKCSYCEKVVSGG-INRFKQHLARIPGEVAPCKSAPEEVYLKIKENMK 195
R+K + C +C K G INR K HLA + A C+ EV ++++++
Sbjct: 41 RRKVITCGFCGKQYVGSEINRMKHHLAGLNNRSA-CQKVSSEVQHSMRKSLE 91
>At1g49590 unknown protein
Length = 242
Score = 35.0 bits (79), Expect = 0.17
Identities = 24/79 (30%), Positives = 34/79 (42%), Gaps = 4/79 (5%)
Query: 197 HRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFK 256
H GKRHR+ K L +S +D E ++ E L + +A R Y KD +
Sbjct: 30 HDLGKRHRECVDKKLTDMRERSAAKDKELKKNEKLLQQIEAKA----TRSYQKDIATAQQ 85
Query: 257 GMSSNTSPEPALRRSRLDS 275
+N +PE LDS
Sbjct: 86 VAKANGAPEDGTSDWMLDS 104
>At3g12640 hypothetical protein
Length = 674
Score = 34.7 bits (78), Expect = 0.23
Identities = 25/100 (25%), Positives = 44/100 (44%), Gaps = 2/100 (2%)
Query: 175 EVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHH 234
E+ P S+ ++ HR KR P+A+ D+ +++ L+
Sbjct: 147 EIPPLLSSEVHKIHNYEKKDHKHRHNKRSPSPQAQSQRKRSRTDDSRNEQRASLRSALYE 206
Query: 235 MNKEALIDIDRRYSKDTGKTFKGMS--SNTSPEPALRRSR 272
NKEA D+ RR + + +S +N+S E +L+R R
Sbjct: 207 TNKEAKPDVSRRLLQFAVRDALAISRPANSSTESSLKRLR 246
>At1g53650 RNA-binding protein, putative
Length = 314
Score = 33.9 bits (76), Expect = 0.39
Identities = 19/68 (27%), Positives = 35/68 (50%), Gaps = 2/68 (2%)
Query: 206 PEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFK--GMSSNTS 263
PEAK+ P Y ++ N+ D++ NK+ I+ RR + + G+ + G +S
Sbjct: 62 PEAKEFFPSYKRNTNQSDDFVIAIKPSGEDNKKVAINRRRRNNYNQGRRVRLPGRASKAQ 121
Query: 264 PEPALRRS 271
E ++RR+
Sbjct: 122 REDSIRRT 129
>At3g45270 putative protein
Length = 661
Score = 33.1 bits (74), Expect = 0.66
Identities = 16/56 (28%), Positives = 25/56 (44%), Gaps = 4/56 (7%)
Query: 14 WDHGIAQDERKKKVRCNYCGKVV----SGGIYRLKQHLARVSGEVTYCEKAPEEVY 65
WDH ++ K+ +CNYC + G LK HL + +KA + V+
Sbjct: 54 WDHYTRFEDNPKRCKCNYCHRTYGCDSKDGTSNLKNHLRICKHYQAWSQKAKQTVF 109
>At1g72440 hypothetical protein
Length = 1056
Score = 33.1 bits (74), Expect = 0.66
Identities = 37/158 (23%), Positives = 65/158 (40%), Gaps = 26/158 (16%)
Query: 113 LMDGRNVSVNLTPLRSLG---YVDPGWE--------HGVAQDERKKKVKCSYCEKVVSGG 161
L+ G N+ N PL L ++D E HG +Q E KK+ S +V+
Sbjct: 787 LLSGTNIVYNGNPLNDLSLTAFLDKFMEKKPKQNTWHGGSQIEPSKKLDMS--NRVIGAE 844
Query: 162 INRFKQHLARIPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDLMPFYPKSDNE 221
I L+ G+VAP + Y+ + K + K+ + PE + Y +D +
Sbjct: 845 I------LSLAEGDVAPEDLVFHKFYVNKMTSTKQSKKKKKKKLPEEEAAEELYDVNDGD 898
Query: 222 -------DDEYEQQEDTLHHMNKEALIDIDRRYSKDTG 252
D E+E +++ + + L D+D ++ G
Sbjct: 899 GGENYDSDVEFEAGDESDNEEIENMLDDVDDNAVEEEG 936
>At5g41270 unknown protein
Length = 258
Score = 31.6 bits (70), Expect = 1.9
Identities = 37/174 (21%), Positives = 72/174 (41%), Gaps = 18/174 (10%)
Query: 149 VKCSYCEKVVSGGIN---RFKQHLARIPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHR- 204
V C CE ++ G N R ++ A + + CK + + + N+ +H HR
Sbjct: 70 VSCQRCETILKPGFNCNVRIEKVSANVKKKRNRCKKSNNICFPQ--NNVVYHCNFCSHRN 127
Query: 205 ------QPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFKGM 258
+ + K+L PF PK+ ++E T+ + ++ R KD +
Sbjct: 128 LKRGTAKGQMKELYPFKPKTARSSRPKIKKEMTMPQEIQSNMLSSPERSVKD---QVEEK 184
Query: 259 SSNTSPEPALRRSRLDSFYLKHPTNQNLQTCKQLKVKT--GPTKKLRKEVFSSI 310
S +P+P + D ++ P ++ + + KT G K+ RK ++S+
Sbjct: 185 SVGDTPKPMMLTLERDR-RIRKPKSKKPSEPQSVPEKTVGGSNKRKRKSPWTSM 237
>At3g28770 hypothetical protein
Length = 2081
Score = 31.6 bits (70), Expect = 1.9
Identities = 37/191 (19%), Positives = 76/191 (39%), Gaps = 8/191 (4%)
Query: 67 KMKENLEGCRSNKKQKQVDAQAYMNFQSNDDEDDEEQVGCRSKGKQLMDGRNVSVNLTPL 126
K K+ E SN K+K+ D + Y+N + ED++++ K + ++
Sbjct: 944 KKKKKKESKNSNMKKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESE 1003
Query: 127 RSLGYVDPGWEH----GVAQDERKKKVKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSA 182
S E+ ++E KK+ K S +K K + K
Sbjct: 1004 DSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKK 1063
Query: 183 PEEVYLKIKENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALID 242
EE K K+ + H++ K+ + E +D K + + E ++ E++ +E D
Sbjct: 1064 EEET--KEKKESENHKSKKKEDKKEHEDNKSM--KKEEDKKEKKKHEESKSRKKEEDKKD 1119
Query: 243 IDRRYSKDTGK 253
+++ +++ K
Sbjct: 1120 MEKLEDQNSNK 1130
>At1g80930 unknown protein
Length = 900
Score = 31.2 bits (69), Expect = 2.5
Identities = 20/76 (26%), Positives = 39/76 (51%), Gaps = 2/76 (2%)
Query: 62 EEVYLKMKENLEGCRSNKKQKQVDAQAYMNFQSND--DEDDEEQVGCRSKGKQLMDGRNV 119
E+ Y +K+ L G ++ + DA + N + D DE+DEEQ+ R + + +
Sbjct: 593 EKKYEALKKELLGDEESEDEDGSDASSEDNDEEEDESDEEDEEQMRIRDETETNLVNLRR 652
Query: 120 SVNLTPLRSLGYVDPG 135
++ LT + S+ + + G
Sbjct: 653 TIYLTIMSSVDFEEAG 668
>At5g48570 peptidylprolyl isomerase
Length = 578
Score = 30.8 bits (68), Expect = 3.3
Identities = 21/83 (25%), Positives = 34/83 (40%)
Query: 58 EKAPEEVYLKMKENLEGCRSNKKQKQVDAQAYMNFQSNDDEDDEEQVGCRSKGKQLMDGR 117
++A YLK+ E +E +S K+K V + N DE + G G + R
Sbjct: 27 DEADSAPYLKIGEEMEIGKSGLKKKLVKECEKWDTPENGDEVEVHYTGTLLDGTKFDSSR 86
Query: 118 NVSVNLTPLRSLGYVDPGWEHGV 140
+ G+V GW+ G+
Sbjct: 87 DRGTPFKFTLGQGHVIKGWDLGI 109
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,206,325
Number of Sequences: 26719
Number of extensions: 848661
Number of successful extensions: 2768
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 2694
Number of HSP's gapped (non-prelim): 61
length of query: 835
length of database: 11,318,596
effective HSP length: 108
effective length of query: 727
effective length of database: 8,432,944
effective search space: 6130750288
effective search space used: 6130750288
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC138579.4


