

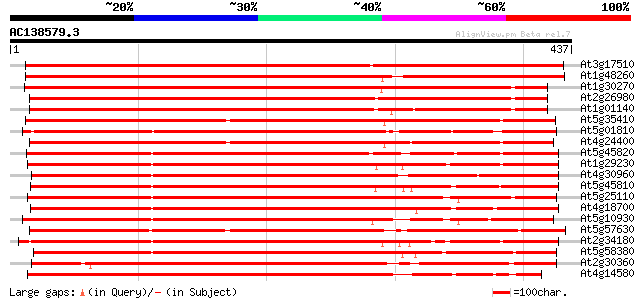
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138579.3 + phase: 0 /pseudo
(437 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g17510 CBL-interacting protein kinase 1 (CIPK1) 590 e-169
At1g48260 CBL-interacting protein kinase 17 (CIPK17) 553 e-158
At1g30270 serine/threonine kinase, putative 450 e-127
At2g26980 putative protein kinase (At2g26980) 429 e-120
At1g01140 SOS2-like protein kinase PKS6/CBL-interacting protein ... 428 e-120
At5g35410 serine/threonine protein kinase SOS2 (gb|AAF62923.1) 402 e-112
At5g01810 serine/threonine protein kinase ATPK10 396 e-110
At4g24400 serine/threonine kinase-like protein 396 e-110
At5g45820 CBL-interacting protein kinase 20 (CIPK20) 376 e-104
At1g29230 CBL-interacting protein kinase 18 (CIPK18) 372 e-103
At4g30960 CBL-interacting protein kinase 6 (CIPK6) 371 e-103
At5g45810 CBL-interacting protein kinase 19 (CIPK19) 369 e-102
At5g25110 serine/threonine protein kinase-like protein 365 e-101
At4g18700 putative protein kinase 365 e-101
At5g10930 serine/threonine protein kinase -like protein 362 e-100
At5g57630 CBL-interacting protein kinase 21 (CIPK21) 362 e-100
At2g34180 putative protein kinase 355 4e-98
At5g58380 SOS2-like protein kinase PKS2 345 2e-95
At2g30360 putative protein kinase 345 3e-95
At4g14580 CBL-interacting protein kinase 4 (CIPK4) 343 8e-95
>At3g17510 CBL-interacting protein kinase 1 (CIPK1)
Length = 444
Score = 590 bits (1521), Expect = e-169
Identities = 301/419 (71%), Positives = 347/419 (81%), Gaps = 1/419 (0%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
K +RLGKYELG+TLGEGNFGKVK A+DT G FAVKI++K++I DLN + QIKREI TL
Sbjct: 13 KGMRLGKYELGRTLGEGNFGKVKFAKDTVSGHSFAVKIIDKSRIADLNFSLQIKREIRTL 72
Query: 73 KLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVS 132
K+LKHP++VRL+EVLASKTKI MV+E V GGELFD+I S GKLTE GRKMFQQLIDG+S
Sbjct: 73 KMLKHPHIVRLHEVLASKTKINMVMELVTGGELFDRIVSNGKLTETDGRKMFQQLIDGIS 132
Query: 133 YCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEIL 192
YCH+KGVFHRDLKLENVL+DAKG+IKITDF LSALPQH R DGLLHTTCGSPNYVAPE+L
Sbjct: 133 YCHSKGVFHRDLKLENVLLDAKGHIKITDFGLSALPQHFRDDGLLHTTCGSPNYVAPEVL 192
Query: 193 ANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNII 252
ANRGYDGA SDIWSCGVILYVILTG LPFDDRNLAVLYQKI K D IPRWLSPGA+ +I
Sbjct: 193 ANRGYDGAASDIWSCGVILYVILTGCLPFDDRNLAVLYQKICKGDPPIPRWLSPGARTMI 252
Query: 253 KKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQS 312
K++LDPNP TR+T+ IK EWFK Y P ++ D+++EE+V DD+AFSI E+ + +
Sbjct: 253 KRMLDPNPVTRITVVGIKASEWFKLEYIP-SIPDDDDEEEVDTDDDAFSIQELGSEEGKG 311
Query: 313 PKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFK 372
SP +INAFQLIGMSS LDLSGFFEQE+VSER+IRFTSN S KDL EKIE V EMGF
Sbjct: 312 SDSPTIINAFQLIGMSSFLDLSGFFEQENVSERRIRFTSNSSAKDLLEKIETAVTEMGFS 371
Query: 373 VHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVCILLL 431
V KK+ KL+V QE++ K LSV AEVFE+ PSL VVELRKSYGD+ +YRQ+ LL
Sbjct: 372 VQKKHAKLRVKQEERNQKGQVGLSVTAEVFEIKPSLNVVELRKSYGDSCLYRQLYERLL 430
>At1g48260 CBL-interacting protein kinase 17 (CIPK17)
Length = 432
Score = 553 bits (1424), Expect = e-158
Identities = 286/424 (67%), Positives = 336/424 (78%), Gaps = 12/424 (2%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
K +R+GKYELG+TLGEGN KVK A DT G+ FA+KI+EK+ I LN + QIKREI TL
Sbjct: 4 KGMRVGKYELGRTLGEGNSAKVKFAIDTLTGESFAIKIIEKSCITRLNVSFQIKREIRTL 63
Query: 73 KLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVS 132
K+LKHPN+VRL+EVLASKTKIYMVLE V GG+LFD+I SKGKL+E GRKMFQQLIDGVS
Sbjct: 64 KVLKHPNIVRLHEVLASKTKIYMVLECVTGGDLFDRIVSKGKLSETQGRKMFQQLIDGVS 123
Query: 133 YCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEIL 192
YCHNKGVFHRDLKLENVL+DAKG+IKITDF LSAL QH R DGLLHTTCGSPNYVAPE+L
Sbjct: 124 YCHNKGVFHRDLKLENVLLDAKGHIKITDFGLSALSQHYREDGLLHTTCGSPNYVAPEVL 183
Query: 193 ANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNII 252
AN GYDGA SDIWSCGVILYVILTG LPFDD NLAV+ +KIFK D IPRW+S GA+ +I
Sbjct: 184 ANEGYDGAASDIWSCGVILYVILTGCLPFDDANLAVICRKIFKGDPPIPRWISLGAKTMI 243
Query: 253 KKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEE----EEDVFIDDEAFSIHEVSLD 308
K++LDPNP TRVT+ IK +WFK Y P+N +D+++ +EDVF+ E +
Sbjct: 244 KRMLDPNPVTRVTIAGIKAHDWFKHDYTPSNYDDDDDVYLIQEDVFMMKE--------YE 295
Query: 309 GDQSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIE 368
++SP SP +INAFQLIGMSS LDLSGFFE E +SER+IRFTSN KDL E IE I E
Sbjct: 296 EEKSPDSPTIINAFQLIGMSSFLDLSGFFETEKLSERQIRFTSNSLAKDLLENIETIFTE 355
Query: 369 MGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVCI 428
MGF + KK+ KLK I+E+ K LSV AEVFE+ PSL VVELRKS+GD+S+Y+Q+
Sbjct: 356 MGFCLQKKHAKLKAIKEESTQKRQCGLSVTAEVFEISPSLNVVELRKSHGDSSLYKQLYE 415
Query: 429 LLLS 432
LL+
Sbjct: 416 RLLN 419
>At1g30270 serine/threonine kinase, putative
Length = 482
Score = 450 bits (1158), Expect = e-127
Identities = 225/412 (54%), Positives = 292/412 (70%), Gaps = 6/412 (1%)
Query: 12 GKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREIST 71
G R+GKYELG+TLGEG F KVK AR+ + G A+K+++K K++ QIKREIST
Sbjct: 23 GGRTRVGKYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMIAQIKREIST 82
Query: 72 LKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGV 131
+KL+KHPNV+R++EV+ASKTKIY VLE+V GGELFDKISS G+L E RK FQQLI+ V
Sbjct: 83 MKLIKHPNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDEARKYFQQLINAV 142
Query: 132 SYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEI 191
YCH++GV+HRDLK EN+L+DA G +K++DF LSALPQ R DGLLHTTCG+PNYVAPE+
Sbjct: 143 DYCHSRGVYHRDLKPENLLLDANGALKVSDFGLSALPQQVREDGLLHTTCGTPNYVAPEV 202
Query: 192 LANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNI 251
+ N+GYDGAK+D+WSCGVIL+V++ GYLPF+D NL LY+KIFKA+ P W S A+ +
Sbjct: 203 INNKGYDGAKADLWSCGVILFVLMAGYLPFEDSNLTSLYKKIFKAEFTCPPWFSASAKKL 262
Query: 252 IKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEE---EEEDVFIDDEAFSIHEVSLD 308
IK+ILDPNP TR+T + E+EWFK+GY E+ + ++ D DD S + V
Sbjct: 263 IKRILDPNPATRITFAEVIENEWFKKGYKAPKFENADVSLDDVDAIFDDSGESKNLVVER 322
Query: 309 GDQSPKSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVI 367
++ K+P +NAF+LI S L+L FE Q + +RK RFTS S ++ KIE
Sbjct: 323 REEGLKTPVTMNAFELISTSQGLNLGSLFEKQMGLVKRKTRFTSKSSANEIVTKIEAAAA 382
Query: 368 EMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
MGF V N K+K+ E K L+VA EVF+V PSLY+VE+RKS GD
Sbjct: 383 PMGFDVKTNNYKMKLTGEKSGRKG--QLAVATEVFQVAPSLYMVEMRKSGGD 432
>At2g26980 putative protein kinase (At2g26980)
Length = 441
Score = 429 bits (1104), Expect = e-120
Identities = 212/404 (52%), Positives = 288/404 (70%), Gaps = 3/404 (0%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+GKYE+G+T+GEG F KVK AR+++ G+ A+KIL+K K++ +QI+REI+T+KL+
Sbjct: 10 RVGKYEVGRTIGEGTFAKVKFARNSETGEPVALKILDKEKVLKHKMAEQIRREIATMKLI 69
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVV+LYEV+ASKTKI+++LEYV GGELFDKI + G++ E R+ FQQLI V YCH
Sbjct: 70 KHPNVVQLYEVMASKTKIFIILEYVTGGELFDKIVNDGRMKEDEARRYFQQLIHAVDYCH 129
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+L+D+ GN+KI+DF LSAL Q R DGLLHT+CG+PNYVAPE+L +R
Sbjct: 130 SRGVYHRDLKPENLLLDSYGNLKISDFGLSALSQQVRDDGLLHTSCGTPNYVAPEVLNDR 189
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGV+LYV+L GYLPFDD NL LY+KI + P WLS GA +I +I
Sbjct: 190 GYDGATADMWSCGVVLYVLLAGYLPFDDSNLMNLYKKISSGEFNCPPWLSLGAMKLITRI 249
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPKS 315
LDPNP TRVT + EDEWFK+ Y P E E ++ ++ D F E L ++ +
Sbjct: 250 LDPNPMTRVTPQEVFEDEWFKKDYKPPVFE-ERDDSNMDDIDAVFKDSEEHLVTEKREEQ 308
Query: 316 PALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKVHK 375
PA INAF++I MS L+L F+ E +R+ R T ++ EKIE+ +GF V K
Sbjct: 309 PAAINAFEIISMSRGLNLENLFDPEQEFKRETRITLRGGANEIIEKIEEAAKPLGFDVQK 368
Query: 376 KNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
KN K+++ K +L+VA E+F+V PSL++V++ KS GD
Sbjct: 369 KNYKMRLENVKAGRKG--NLNVATEIFQVAPSLHMVQVSKSKGD 410
>At1g01140 SOS2-like protein kinase PKS6/CBL-interacting protein
kinase 9 (CIPK9)
Length = 447
Score = 428 bits (1101), Expect = e-120
Identities = 214/407 (52%), Positives = 294/407 (71%), Gaps = 8/407 (1%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+G YE+G+TLGEG+F KVK A++T G A+KIL++ K+ +Q+KREIST+KL+
Sbjct: 15 RVGNYEMGRTLGEGSFAKVKYAKNTVTGDQAAIKILDREKVFRHKMVEQLKREISTMKLI 74
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVV + EV+ASKTKIY+VLE VNGGELFDKI+ +G+L E R+ FQQLI+ V YCH
Sbjct: 75 KHPNVVEIIEVMASKTKIYIVLELVNGGELFDKIAQQGRLKEDEARRYFQQLINAVDYCH 134
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+++DA G +K++DF LSA + R DGLLHT CG+PNYVAPE+L+++
Sbjct: 135 SRGVYHRDLKPENLILDANGVLKVSDFGLSAFSRQVREDGLLHTACGTPNYVAPEVLSDK 194
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGVIL+V++ GYLPFD+ NL LY++I KA+ P W S GA+ +IK+I
Sbjct: 195 GYDGAAADVWSCGVILFVLMAGYLPFDEPNLMTLYKRICKAEFSCPPWFSQGAKRVIKRI 254
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFID--DEAFSIHEVSLDGDQSP 313
L+PNP TR+++ + EDEWFK+GY P + ++++ED+ ID D AFS + L ++
Sbjct: 255 LEPNPITRISIAELLEDEWFKKGYKPPSF--DQDDEDITIDDVDAAFSNSKECLVTEKKE 312
Query: 314 KSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFK 372
K P +NAF+LI SS L FE Q + +++ RFTS S ++ K+E+ +GF
Sbjct: 313 K-PVSMNAFELISSSSEFSLENLFEKQAQLVKKETRFTSQRSASEIMSKMEETAKPLGFN 371
Query: 373 VHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
V K N K+K+ + K LSVA EVFEV PSL+VVELRK+ GD
Sbjct: 372 VRKDNYKIKMKGDKSGRKG--QLSVATEVFEVAPSLHVVELRKTGGD 416
>At5g35410 serine/threonine protein kinase SOS2 (gb|AAF62923.1)
Length = 446
Score = 402 bits (1033), Expect = e-112
Identities = 202/416 (48%), Positives = 288/416 (68%), Gaps = 7/416 (1%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
K+ R+GKYE+G+T+GEG F KVK AR+TD G A+KI+ K+ I+ DQIKREIS +
Sbjct: 4 KMRRVGKYEVGRTIGEGTFAKVKFARNTDTGDNVAIKIMAKSTILKNRMVDQIKREISIM 63
Query: 73 KLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVS 132
K+++HPN+VRLYEVLAS +KIY+VLE+V GGELFD+I KG+L E+ RK FQQL+D V+
Sbjct: 64 KIVRHPNIVRLYEVLASPSKIYIVLEFVTGGELFDRIVHKGRLEESESRKYFQQLVDAVA 123
Query: 133 YCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEIL 192
+CH KGV+HRDLK EN+L+D GN+K++DF LSALPQ LL TTCG+PNYVAPE+L
Sbjct: 124 HCHCKGVYHRDLKPENLLLDTNGNLKVSDFGLSALPQ--EGVELLRTTCGTPNYVAPEVL 181
Query: 193 ANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNII 252
+ +GYDG+ +DIWSCGVIL+VIL GYLPF + +L LY+KI A+ P W S + +I
Sbjct: 182 SGQGYDGSAADIWSCGVILFVILAGYLPFSETDLPGLYRKINAAEFSCPPWFSAEVKFLI 241
Query: 253 KKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEE--EDVFIDDEAFSIHEVSLDGD 310
+ILDPNPKTR+ + IK+D WF+ Y P +EEE +D+ + V+ + +
Sbjct: 242 HRILDPNPKTRIQIQGIKKDPWFRLNYVPIRAREEEEVNLDDIRAVFDGIEGSYVAENVE 301
Query: 311 QSPKSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVIEM 369
++ + P ++NAF++I +S L+LS F+ ++D +R+ RF S P ++ IE + M
Sbjct: 302 RNDEGPLMMNAFEMITLSQGLNLSALFDRRQDFVKRQTRFVSRREPSEIIANIEAVANSM 361
Query: 370 GFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQ 425
GFK H +N K ++ E + L+V E++EV PSL++V++RK+ G+ Y +
Sbjct: 362 GFKSHTRNFKTRL--EGLSSIKAGQLAVVIEIYEVAPSLFMVDVRKAAGETLEYHK 415
>At5g01810 serine/threonine protein kinase ATPK10
Length = 421
Score = 396 bits (1018), Expect = e-110
Identities = 204/416 (49%), Positives = 289/416 (69%), Gaps = 15/416 (3%)
Query: 11 EGKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREIS 70
+G ++ L +YE+GK LG+G F KV AR G A+K+++K +I+ + T+QIKREIS
Sbjct: 4 KGSVLML-RYEVGKFLGQGTFAKVYHARHLKTGDSVAIKVIDKERILKVGMTEQIKREIS 62
Query: 71 TLKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDG 130
++LL+HPN+V L+EV+A+K+KIY V+E+V GGELF+K+S+ GKL E RK FQQL+
Sbjct: 63 AMRLLRHPNIVELHEVMATKSKIYFVMEHVKGGELFNKVST-GKLREDVARKYFQQLVRA 121
Query: 131 VSYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPE 190
V +CH++GV HRDLK EN+L+D GN+KI+DF LSAL R DGLLHTTCG+P YVAPE
Sbjct: 122 VDFCHSRGVCHRDLKPENLLLDEHGNLKISDFGLSALSDSRRQDGLLHTTCGTPAYVAPE 181
Query: 191 ILANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQN 250
+++ GYDG K+D+WSCGVIL+V+L GYLPF D NL LY+KI KA+V+ P WL+PGA+
Sbjct: 182 VISRNGYDGFKADVWSCGVILFVLLAGYLPFRDSNLMELYKKIGKAEVKFPNWLAPGAKR 241
Query: 251 IIKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGD 310
++K+ILDPNP TRV+ + I + WF++G E EEE +V D EA E + +
Sbjct: 242 LLKRILDPNPNTRVSTEKIMKSSWFRKGLQEEVKESVEEETEV--DAEA----EGNASAE 295
Query: 311 QSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMG 370
+ K +NAF++I +S+ DLSG FE+ + E ++RFTSN ++TEK+ +I ++
Sbjct: 296 KEKKRCINLNAFEIISLSTGFDLSGLFEKGEEKE-EMRFTSNREASEITEKLVEIGKDLK 354
Query: 371 FKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQV 426
KV KK E + S ++ V AEVFE+ PS ++V L+KS GD + Y++V
Sbjct: 355 MKVRKKE------HEWRVKMSAEATVVEAEVFEIAPSYHMVVLKKSGGDTAEYKRV 404
>At4g24400 serine/threonine kinase-like protein
Length = 445
Score = 396 bits (1017), Expect = e-110
Identities = 201/411 (48%), Positives = 289/411 (69%), Gaps = 8/411 (1%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
++GKYELG+T+GEG F KVK A++T+ G+ A+KI++++ I+ DQIKREIS +KL+
Sbjct: 5 KVGKYELGRTIGEGTFAKVKFAQNTETGESVAMKIVDRSTIIKRKMVDQIKREISIMKLV 64
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
+HP VVRLYEVLAS+TKIY++LEY+ GGELFDKI G+L+E+ RK F QLIDGV YCH
Sbjct: 65 RHPCVVRLYEVLASRTKIYIILEYITGGELFDKIVRNGRLSESEARKYFHQLIDGVDYCH 124
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
+KGV+HRDLK EN+L+D++GN+KI+DF LSALP+ + +L TTCG+PNYVAPE+L+++
Sbjct: 125 SKGVYHRDLKPENLLLDSQGNLKISDFGLSALPE--QGVTILKTTCGTPNYVAPEVLSHK 182
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GY+GA +DIWSCGVILYV++ GYLPFD+ +L LY KI KA+ P + + GA+++I +I
Sbjct: 183 GYNGAVADIWSCGVILYVLMAGYLPFDEMDLPTLYSKIDKAEFSCPSYFALGAKSLINRI 242
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEE--EDVFIDDEAFSIHEVSLDGDQSP 313
LDPNP+TR+T+ I++DEWF + Y P L D E +DV+ + + DG +
Sbjct: 243 LDPNPETRITIAEIRKDEWFLKDYTPVQLIDYEHVNLDDVYAAFDDPEEQTYAQDGTRD- 301
Query: 314 KSPALINAFQLIGMSSCLDLSGFFEQ-EDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFK 372
P +NAF LI +S L+L+ F++ +D + + RF S+ + +E + MGFK
Sbjct: 302 TGPLTLNAFDLIILSQGLNLATLFDRGKDSMKHQTRFISHKPANVVLSSMEVVSQSMGFK 361
Query: 373 VHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVY 423
H +N K++V E + SV EVF+V PS+ +V+++ + GDA Y
Sbjct: 362 THIRNYKMRV--EGLSANKTSHFSVILEVFKVAPSILMVDIQNAAGDAEEY 410
>At5g45820 CBL-interacting protein kinase 20 (CIPK20)
Length = 439
Score = 376 bits (966), Expect = e-104
Identities = 194/416 (46%), Positives = 273/416 (64%), Gaps = 15/416 (3%)
Query: 14 IVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLK 73
IV + KYELG+ LG+G F KV AR+ G+ A+K+++K K+ + DQIKREIS ++
Sbjct: 6 IVLMRKYELGRLLGQGTFAKVYHARNIKTGESVAIKVIDKQKVAKVGLIDQIKREISVMR 65
Query: 74 LLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSY 133
L++HP+VV L+EV+ASKTKIY +EYV GGELFDK+S KGKL E RK FQQLI + Y
Sbjct: 66 LVRHPHVVFLHEVMASKTKIYFAMEYVKGGELFDKVS-KGKLKENIARKYFQQLIGAIDY 124
Query: 134 CHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILA 193
CH++GV+HRDLK EN+L+D G++KI+DF LSAL + + DGLLHTTCG+P YVAPE++
Sbjct: 125 CHSRGVYHRDLKPENLLLDENGDLKISDFGLSALRESKQQDGLLHTTCGTPAYVAPEVIG 184
Query: 194 NRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIK 253
+GYDGAK+D+WSCGV+LYV+L G+LPF ++NL +Y+KI K + + P W P + ++
Sbjct: 185 KKGYDGAKADVWSCGVVLYVLLAGFLPFHEQNLVEMYRKITKGEFKCPNWFPPEVKKLLS 244
Query: 254 KILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSP 313
+ILDPNP +R+ ++ I E+ WF++G+ +E + E ID +H
Sbjct: 245 RILDPNPNSRIKIEKIMENSWFQKGFK--KIETPKSPESHQIDSLISDVHAA------FS 296
Query: 314 KSPALINAFQLI-GMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEM-GF 371
P NAF LI +S DLSG FE+E+ SE K FT+ K++ K E+I F
Sbjct: 297 VKPMSYNAFDLISSLSQGFDLSGLFEKEERSESK--FTTKKDAKEIVSKFEEIATSSERF 354
Query: 372 KVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVC 427
+ K + +K+ EDK L++ E+FEV S ++VE +KS GD Y+Q C
Sbjct: 355 NLTKSDVGVKM--EDKREGRKGHLAIDVEIFEVTNSFHMVEFKKSGGDTMEYKQFC 408
>At1g29230 CBL-interacting protein kinase 18 (CIPK18)
Length = 520
Score = 372 bits (954), Expect = e-103
Identities = 189/428 (44%), Positives = 273/428 (63%), Gaps = 20/428 (4%)
Query: 15 VRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKL 74
+ +GKYELGK LG G F KV LA++ G A+K+++K KI+ IKREIS L+
Sbjct: 69 ILMGKYELGKLLGHGTFAKVYLAQNIKSGDKVAIKVIDKEKIMKSGLVAHIKREISILRR 128
Query: 75 LKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYC 134
++HP +V L+EV+A+K+KIY V+EYV GGELF+ ++ KG+L E R+ FQQLI VS+C
Sbjct: 129 VRHPYIVHLFEVMATKSKIYFVMEYVGGGELFNTVA-KGRLPEETARRYFQQLISSVSFC 187
Query: 135 HNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILAN 194
H +GV+HRDLK EN+L+D KGN+K++DF LSA+ + R DGL HT CG+P Y+APE+L
Sbjct: 188 HGRGVYHRDLKPENLLLDNKGNLKVSDFGLSAVAEQLRQDGLCHTFCGTPAYIAPEVLTR 247
Query: 195 RGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKK 254
+GYD AK+D+WSCGVIL+V++ G++PF D+N+ V+Y+KI+K + + PRW S ++ +
Sbjct: 248 KGYDAAKADVWSCGVILFVLMAGHIPFYDKNIMVMYKKIYKGEFRCPRWFSSDLVRLLTR 307
Query: 255 ILDPNPKTRVTMDMIKEDEWFKEGYNPANL---------EDEEEEEDVFIDDEAFSIHE- 304
+LD NP TR+T+ I ++ WFK+G+ EDE+EEE+ + ++ E
Sbjct: 308 LLDTNPDTRITIPEIMKNRWFKKGFKHVKFYIEDDKLCREDEDEEEEASSSGRSSTVSES 367
Query: 305 -----VSLDGDQSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLT 359
V G S P+ +NAF +I SS DLSG FE+E RF S +
Sbjct: 368 DAEFDVKRMGIGSMPRPSSLNAFDIISFSSGFDLSGLFEEE--GGEGTRFVSGAPVSKII 425
Query: 360 EKIEDIVIEMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
K+E+I + F V KK L++ E + L++AAE+FE+ PSL VVE++K GD
Sbjct: 426 SKLEEIAKIVSFTVRKKEWSLRL--EGCREGAKGPLTIAAEIFELTPSLVVVEVKKKGGD 483
Query: 420 ASVYRQVC 427
Y + C
Sbjct: 484 REEYEEFC 491
>At4g30960 CBL-interacting protein kinase 6 (CIPK6)
Length = 441
Score = 371 bits (953), Expect = e-103
Identities = 184/411 (44%), Positives = 266/411 (63%), Gaps = 10/411 (2%)
Query: 18 GKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLLKH 77
G+YELG+ LG G F KV AR+ G+ A+K++ K K+V + DQIKREIS ++++KH
Sbjct: 22 GRYELGRLLGHGTFAKVYHARNIQTGKSVAMKVVGKEKVVKVGMVDQIKREISVMRMVKH 81
Query: 78 PNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCHNK 137
PN+V L+EV+ASK+KIY +E V GGELF K++ KG+L E R FQQLI V +CH++
Sbjct: 82 PNIVELHEVMASKSKIYFAMELVRGGELFAKVA-KGRLREDVARVYFQQLISAVDFCHSR 140
Query: 138 GVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANRGY 197
GV+HRDLK EN+L+D +GN+K+TDF LSA +H + DGLLHTTCG+P YVAPE++ +GY
Sbjct: 141 GVYHRDLKPENLLLDEEGNLKVTDFGLSAFTEHLKQDGLLHTTCGTPAYVAPEVILKKGY 200
Query: 198 DGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKILD 257
DGAK+D+WSCGVIL+V+L GYLPF D NL +Y+KI++ D + P WLS A+ ++ K+LD
Sbjct: 201 DGAKADLWSCGVILFVLLAGYLPFQDDNLVNMYRKIYRGDFKCPGWLSSDARRLVTKLLD 260
Query: 258 PNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPKSPA 317
PNP TR+T++ + + WFK+ + E +D F +H +S +
Sbjct: 261 PNPNTRITIEKVMDSPWFKKQATRSRNEPVAATITTTEEDVDFLVH-------KSKEETE 313
Query: 318 LINAFQLIGMSSCLDLSGFFEQEDVSE-RKIRFTSNHSPKDLTEKIEDIVIEMGFKVHKK 376
+NAF +I +S DLS FE++ E R++RF ++ + +E+ +G K +
Sbjct: 314 TLNAFHIIALSEGFDLSPLFEEKKKEEKREMRFATSRPASSVISSLEE-AARVGNKFDVR 372
Query: 377 NGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVC 427
+ +V E K + L+V AE+F V PS VVE++K +GD Y C
Sbjct: 373 KSESRVRIEGKQNGRKGKLAVEAEIFAVAPSFVVVEVKKDHGDTLEYNNFC 423
>At5g45810 CBL-interacting protein kinase 19 (CIPK19)
Length = 483
Score = 369 bits (948), Expect = e-102
Identities = 195/430 (45%), Positives = 272/430 (62%), Gaps = 24/430 (5%)
Query: 17 LGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLLK 76
LGKYE+G+ LG G F KV LAR+ G+ A+K+++K K++ IKREIS L+ ++
Sbjct: 25 LGKYEMGRLLGHGTFAKVYLARNAQSGESVAIKVIDKEKVLKSGLIAHIKREISILRRVR 84
Query: 77 HPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCHN 136
HPN+V+L+EV+A+K+KIY V+EYV GGELF+K++ KG+L E RK FQQLI VS+CH
Sbjct: 85 HPNIVQLFEVMATKSKIYFVMEYVKGGELFNKVA-KGRLKEEMARKYFQQLISAVSFCHF 143
Query: 137 KGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANRG 196
+GV+HRDLK EN+L+D GN+K++DF LSA+ R DGL HT CG+P YVAPE+LA +G
Sbjct: 144 RGVYHRDLKPENLLLDENGNLKVSDFGLSAVSDQIRQDGLFHTFCGTPAYVAPEVLARKG 203
Query: 197 YDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKIL 256
YDGAK DIWSCGVIL+V++ G+LPF DRN+ +Y+KI++ D + PRW ++ ++L
Sbjct: 204 YDGAKVDIWSCGVILFVLMAGFLPFHDRNVMAMYKKIYRGDFRCPRWFPVEINRLLIRML 263
Query: 257 DPNPKTRVTMDMIKEDEWFKEGYNPAN--LEDEEEEEDVFIDDEAFSIHEV--------- 305
+ P+ R TM I E WFK+G+ +ED+ + +V DDE SI V
Sbjct: 264 ETKPERRFTMPDIMETSWFKKGFKHIKFYVEDDHQLCNVADDDEIESIESVSGRSSTVSE 323
Query: 306 -----SLDGDQ---SPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKD 357
S DG + S PA +NAF LI S DLSG FE + RF S
Sbjct: 324 PEDFESFDGRRRGGSMPRPASLNAFDLISFSPGFDLSGLFEDDGEGS---RFVSGAPVGQ 380
Query: 358 LTEKIEDIVIEMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSY 417
+ K+E+I + F V KK+ K+ ++ + LS+AAE+FE+ P+L VVE++K
Sbjct: 381 IISKLEEIARIVSFTVRKKDCKVS-LEGSREGSMKGPLSIAAEIFELTPALVVVEVKKKG 439
Query: 418 GDASVYRQVC 427
GD Y + C
Sbjct: 440 GDKMEYDEFC 449
>At5g25110 serine/threonine protein kinase-like protein
Length = 488
Score = 365 bits (938), Expect = e-101
Identities = 195/416 (46%), Positives = 270/416 (64%), Gaps = 11/416 (2%)
Query: 15 VRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKL 74
V KYE+G+ LG+G FGKV ++ G+ A+KI+ K+++ +QIKREIS ++L
Sbjct: 38 VLFAKYEMGRLLGKGTFGKVYYGKEITTGESVAIKIINKDQVKREGMMEQIKREISIMRL 97
Query: 75 LKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYC 134
++HPN+V L EV+A+KTKI+ ++EYV GGELF KI KGKL E RK FQQLI V +C
Sbjct: 98 VRHPNIVELKEVMATKTKIFFIMEYVKGGELFSKIV-KGKLKEDSARKYFQQLISAVDFC 156
Query: 135 HNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILAN 194
H++GV HRDLK EN+LVD G++K++DF LSALP+ DGLLHT CG+P YVAPE+L
Sbjct: 157 HSRGVSHRDLKPENLLVDENGDLKVSDFGLSALPEQILQDGLLHTQCGTPAYVAPEVLRK 216
Query: 195 RGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKK 254
+GYDGAK DIWSCG+ILYV+L G+LPF D NL +Y+KIFK++ + P W SP ++ +I K
Sbjct: 217 KGYDGAKGDIWSCGIILYVLLAGFLPFQDENLMKMYRKIFKSEFEYPPWFSPESKRLISK 276
Query: 255 ILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPK 314
+L +P R+++ I WF++ N +E E ++DE + + +P
Sbjct: 277 LLVVDPNKRISIPAIMRTPWFRKNINSPIEFKIDELEIQNVEDETPTTTATTATTTTTPV 336
Query: 315 SPALINAFQLI-GMSSCLDLSGFFEQEDVSERKIR--FTSNHSPKDLTEKIEDIVIEMGF 371
SP NAF+ I MSS DLS FE S+RK+R FTS S ++ K+E I EM
Sbjct: 337 SPKFFNAFEFISSMSSGFDLSSLFE----SKRKLRSMFTSRWSASEIMGKLEGIGKEMNM 392
Query: 372 KVHK-KNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQV 426
KV + K+ K+K+ + + K ++V AEVFEV P + VVEL KS GD Y ++
Sbjct: 393 KVKRTKDFKVKLFGKTEGRKG--QIAVTAEVFEVAPEVAVVELCKSAGDTLEYNRL 446
>At4g18700 putative protein kinase
Length = 489
Score = 365 bits (936), Expect = e-101
Identities = 186/428 (43%), Positives = 263/428 (60%), Gaps = 23/428 (5%)
Query: 17 LGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLLK 76
LG+YE+GK LG G F KV LAR+ + A+K+++K K++ IKREIS L+ ++
Sbjct: 23 LGRYEMGKLLGHGTFAKVYLARNVKTNESVAIKVIDKEKVLKGGLIAHIKREISILRRVR 82
Query: 77 HPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCHN 136
HPN+V+L+EV+A+K KIY V+EYV GGELF+K++ KG+L E RK FQQLI V++CH
Sbjct: 83 HPNIVQLFEVMATKAKIYFVMEYVRGGELFNKVA-KGRLKEEVARKYFQQLISAVTFCHA 141
Query: 137 KGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANRG 196
+GV+HRDLK EN+L+D GN+K++DF LSA+ R DGL HT CG+P YVAPE+LA +G
Sbjct: 142 RGVYHRDLKPENLLLDENGNLKVSDFGLSAVSDQIRQDGLFHTFCGTPAYVAPEVLARKG 201
Query: 197 YDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKIL 256
YD AK DIWSCGVIL+V++ GYLPF DRN+ +Y+KI++ + + PRW S ++ K+L
Sbjct: 202 YDAAKVDIWSCGVILFVLMAGYLPFHDRNVMAMYKKIYRGEFRCPRWFSTELTRLLSKLL 261
Query: 257 DPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPKS- 315
+ NP+ R T I E+ WFK+G+ E+++ +DD+ V D D +
Sbjct: 262 ETNPEKRFTFPEIMENSWFKKGFKHIKFYVEDDKLCNVVDDDELESDSVESDRDSAASES 321
Query: 316 ----------------PALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLT 359
PA +NAF +I S DLSG F+ + RF S +
Sbjct: 322 EIEYLEPRRRVGGLPRPASLNAFDIISFSQGFDLSGLFDDDGEGS---RFVSGAPVSKII 378
Query: 360 EKIEDIVIEMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
K+E+I + F V KK+ +V E L++AAE+FE+ PSL VVE++K GD
Sbjct: 379 SKLEEIAKVVSFTVRKKD--CRVSLEGSRQGVKGPLTIAAEIFELTPSLVVVEVKKKGGD 436
Query: 420 ASVYRQVC 427
+ Y C
Sbjct: 437 KTEYEDFC 444
>At5g10930 serine/threonine protein kinase -like protein
Length = 445
Score = 362 bits (929), Expect = e-100
Identities = 201/430 (46%), Positives = 270/430 (62%), Gaps = 35/430 (8%)
Query: 11 EGKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNT-DQIKREI 69
E + V GKYE+G+ LG+G F KV ++ G+ A+K++ K++++ +QIKREI
Sbjct: 3 EERRVLFGKYEMGRLLGKGTFAKVYYGKEIIGGECVAIKVINKDQVMKRPGMMEQIKREI 62
Query: 70 STLKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLID 129
S +KL++HPN+V L EV+A+KTKI+ V+E+V GGELF KIS KGKL E R+ FQQLI
Sbjct: 63 SIMKLVRHPNIVELKEVMATKTKIFFVMEFVKGGELFCKIS-KGKLHEDAARRYFQQLIS 121
Query: 130 GVSYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAP 189
V YCH++GV HRDLK EN+L+D G++KI+DF LSALP+ DGLLHT CG+P YVAP
Sbjct: 122 AVDYCHSRGVSHRDLKPENLLLDENGDLKISDFGLSALPEQILQDGLLHTQCGTPAYVAP 181
Query: 190 EILANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQ 249
E+L +GYDGAK+DIWSCGV+LYV+L G LPF D NL +Y+KIF+AD + P W SP A+
Sbjct: 182 EVLKKKGYDGAKADIWSCGVVLYVLLAGCLPFQDENLMNMYRKIFRADFEFPPWFSPEAR 241
Query: 250 NIIKKILDPNPKTRVTMDMIKEDEWFKEGYNP-------------ANLEDEEEEEDVFID 296
+I K+L +P R+++ I W ++ + P ++ +EEEEED +
Sbjct: 242 RLISKLLVVDPDRRISIPAIMRTPWLRKNFTPPLAFKIDEPICSQSSKNNEEEEEDGDCE 301
Query: 297 DEAFSIHEVSLDGDQSPKSPALINAFQLI-GMSSCLDLSGFFEQEDVSERKIR--FTSNH 353
++ P SP NAF+ I MSS DLS FE S+RK++ FTS
Sbjct: 302 NQT------------EPISPKFFNAFEFISSMSSGFDLSSLFE----SKRKVQSVFTSRS 345
Query: 354 SPKDLTEKIEDIVIEMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVEL 413
S ++ EKIE + EM KV K+ KV E K LS+ AEVFEV P + VVE
Sbjct: 346 SATEVMEKIETVTKEMNMKV-KRTKDFKVKMEGKTEGRKGRLSMTAEVFEVAPEISVVEF 404
Query: 414 RKSYGDASVY 423
KS GD Y
Sbjct: 405 CKSAGDTLEY 414
>At5g57630 CBL-interacting protein kinase 21 (CIPK21)
Length = 416
Score = 362 bits (928), Expect = e-100
Identities = 192/421 (45%), Positives = 280/421 (65%), Gaps = 21/421 (4%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
++GKYE+G+T+GEGNF KVKL DT G + AVKI++K ++ Q+KREI T+KLL
Sbjct: 8 KIGKYEIGRTIGEGNFAKVKLGYDTTNGTYVAVKIIDKALVIQKGLESQVKREIRTMKLL 67
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
HPN+V+++EV+ +KTKI +V+EYV+GG+L D++ + K+ E+ RK+FQQLID V YCH
Sbjct: 68 NHPNIVQIHEVIGTKTKICIVMEYVSGGQLSDRL-GRQKMKESDARKLFQQLIDAVDYCH 126
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
N+GV+HRDLK +N+L+D+KGN+K++DF LSA+P ++ +L T CGSP Y+APE++ N+
Sbjct: 127 NRGVYHRDLKPQNLLLDSKGNLKVSDFGLSAVP---KSGDMLSTACGSPCYIAPELIMNK 183
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GY GA D+WSCGVIL+ +L GY PFDD L VLY+KI +AD P + + +I I
Sbjct: 184 GYSGAAVDVWSCGVILFELLAGYPPFDDHTLPVLYKKILRADYTFPPGFTGEQKRLIFNI 243
Query: 256 LDPNPKTRVTM-DMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPK 314
LDPNP +R+T+ ++I +D WFK GY P + + +D ++ E+ + +
Sbjct: 244 LDPNPLSRITLAEIIIKDSWFKIGYTPVYHQLSDSIKD--------NVAEI----NAATA 291
Query: 315 SPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKVH 374
S INAFQ+I MSS LDLSG FE+ D K R S ++ ++ +KIE + V
Sbjct: 292 SSNFINAFQIIAMSSDLDLSGLFEENDDKRYKTRIGSKNTAQETIKKIEAAATYVSLSVE 351
Query: 375 K-KNGKLKV-IQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVCILLLS 432
+ K+ K+K+ +E ++ S D LS AEV EV P+ V+E+ KS G+ +Y + C L S
Sbjct: 352 RIKHFKVKIQPKEIRSRSSYDLLS--AEVIEVTPTNCVIEISKSAGELRLYMEFCQSLSS 409
Query: 433 L 433
L
Sbjct: 410 L 410
>At2g34180 putative protein kinase
Length = 502
Score = 355 bits (910), Expect = 4e-98
Identities = 184/436 (42%), Positives = 278/436 (63%), Gaps = 26/436 (5%)
Query: 8 RRIEGKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKR 67
R +G I+ + KYE+GK LG G+F KV LAR+ G+ A+K+++K KIV IKR
Sbjct: 46 RTPQGSIL-MDKYEIGKLLGHGSFAKVYLARNIHSGEDVAIKVIDKEKIVKSGLAGHIKR 104
Query: 68 EISTLKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQL 127
EIS L+ ++HP +V L EV+A+KTKIY+V+EYV GGEL++ ++ +G+L E R+ FQQL
Sbjct: 105 EISILRRVRHPYIVHLLEVMATKTKIYIVMEYVRGGELYNTVA-RGRLREGTARRYFQQL 163
Query: 128 IDGVSYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYV 187
I V++CH++GV+HRDLKLEN+L+D KGN+K++DF LS + + + +G+ T CG+P Y+
Sbjct: 164 ISSVAFCHSRGVYHRDLKLENLLLDDKGNVKVSDFGLSVVSEQLKQEGICQTFCGTPAYL 223
Query: 188 APEILANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPG 247
APE+L +GY+GAK+DIWSCGVIL+V++ GYLPFDD+N+ V+Y KI+K + P+W SP
Sbjct: 224 APEVLTRKGYEGAKADIWSCGVILFVLMAGYLPFDDKNILVMYTKIYKGQFKCPKWFSPE 283
Query: 248 AQNIIKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEE----EEDVFIDDEAFSI- 302
++ ++LD NP TR+T+ I + WFK+G+ E + E+D DD++ S+
Sbjct: 284 LARLVTRMLDTNPDTRITIPEIMKHRWFKKGFKHVKFYIENDKLCREDDDNDDDDSSSLS 343
Query: 303 ---HEVSLDGD--------QSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTS 351
+ +GD S PA +NAF ++ S DLSG FE+ + RF S
Sbjct: 344 SGRSSTASEGDAEFDIKRVDSMPRPASLNAFDILSFS---DLSGLFEE---GGQGARFVS 397
Query: 352 NHSPKDLTEKIEDIVIEMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVV 411
+ K+E+I E+ F V KK+ +++ E + L++ E+FE+ PSL VV
Sbjct: 398 AAPMTKIISKLEEIAKEVKFMVRKKDWSVRL--EGCREGAKGPLTIRVEIFELTPSLVVV 455
Query: 412 ELRKSYGDASVYRQVC 427
E++K G+ Y + C
Sbjct: 456 EVKKKGGNIEEYEEFC 471
>At5g58380 SOS2-like protein kinase PKS2
Length = 479
Score = 345 bits (886), Expect = 2e-95
Identities = 178/426 (41%), Positives = 276/426 (64%), Gaps = 23/426 (5%)
Query: 19 KYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLLKHP 78
KY++G+ LG+G F KV R Q A+K+++K K++ + +QIKREIS +++ +HP
Sbjct: 11 KYDVGRLLGQGTFAKVYYGRSILTNQSVAIKMIDKEKVMKVGLIEQIKREISVMRIARHP 70
Query: 79 NVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCHNKG 138
NVV LYEV+A+KT+IY V+EY GGELF+K++ KGKL + K F QLI+ V +CH++
Sbjct: 71 NVVELYEVMATKTRIYFVMEYCKGGELFNKVA-KGKLRDDVAWKYFYQLINAVDFCHSRE 129
Query: 139 VFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANRGYD 198
V+HRD+K EN+L+D N+K++DF LSAL R DGLLHTTCG+P YVAPE++ +GYD
Sbjct: 130 VYHRDIKPENLLLDDNENLKVSDFGLSALADCKRQDGLLHTTCGTPAYVAPEVINRKGYD 189
Query: 199 GAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKILDP 258
G K+DIWSCGV+L+V+L GYLPF D NL +Y+KI KAD + P W +P + ++ K+LDP
Sbjct: 190 GTKADIWSCGVVLFVLLAGYLPFHDSNLMEMYRKIGKADFKAPSWFAPEVRRLLCKMLDP 249
Query: 259 NPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDV-FIDDEAFSIHE-----------VS 306
NP+TR+T+ I+E WF++G + + E+ +++ ++ +E S
Sbjct: 250 NPETRITIARIRESSWFRKGLHMKQKKMEKRVKEINSVEAGTAGTNENGAGPSENGAGPS 309
Query: 307 LDGDQSPK-----SPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEK 361
+GD+ + P +NAF LI +S+ DL+G F D ++R+ RFTS + K
Sbjct: 310 ENGDRVTEENHTDEPTNLNAFDLIALSAGFDLAGLF--GDDNKRESRFTSQKPASVIISK 367
Query: 362 IEDIVIEMGFKVHKKN-GKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDA 420
+E++ + + K+ G K+ + + K + LS+ AE+F+V P+ ++VE++KS GD
Sbjct: 368 LEEVAQRLKLSIRKREAGLFKLERLKEGRKGI--LSMDAEIFQVTPNFHLVEVKKSNGDT 425
Query: 421 SVYRQV 426
Y+++
Sbjct: 426 LEYQKL 431
>At2g30360 putative protein kinase
Length = 435
Score = 345 bits (885), Expect = 3e-95
Identities = 186/414 (44%), Positives = 260/414 (61%), Gaps = 25/414 (6%)
Query: 18 GKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNN---TDQIKREISTLKL 74
GKYELGK LG G F KV ARD GQ AVKIL K K+ L N + IKREIS ++
Sbjct: 19 GKYELGKLLGCGAFAKVFHARDRRTGQSVAVKILNKKKL--LTNPALANNIKREISIMRR 76
Query: 75 LKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYC 134
L HPN+V+L+EV+A+K+KI+ +E+V GGELF+KIS G+L+E R+ FQQLI V YC
Sbjct: 77 LSHPNIVKLHEVMATKSKIFFAMEFVKGGELFNKISKHGRLSEDLSRRYFQQLISAVGYC 136
Query: 135 HNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILAN 194
H +GV+HRDLK EN+L+D GN+K++DF LSAL R DGLLHT CG+P YVAPEIL+
Sbjct: 137 HARGVYHRDLKPENLLIDENGNLKVSDFGLSALTDQIRPDGLLHTLCGTPAYVAPEILSK 196
Query: 195 RGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKK 254
+GY+GAK D+WSCG++L+V++ GYLPF+D N+ +Y+KI+K + + PRW+SP + + +
Sbjct: 197 KGYEGAKVDVWSCGIVLFVLVAGYLPFNDPNVMNMYKKIYKGEYRFPRWMSPDLKRFVSR 256
Query: 255 ILDPNPKTRVTMDMIKEDEWF-KEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSP 313
+LD NP+TR+T+D I +D WF + G+ D+E E+ E SL+ +S
Sbjct: 257 LLDINPETRITIDEILKDPWFVRGGFKQIKFHDDEIEDQKV---------ESSLEAVKS- 306
Query: 314 KSPALINAFQLIGMSSCLDLSGFFEQ-EDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFK 372
+NAF LI SS LDLSG F + S RF S SP+ L E++E E +
Sbjct: 307 -----LNAFDLISYSSGLDLSGLFAGCSNSSGESERFLSEKSPEMLAEEVEGFAREENLR 361
Query: 373 VHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQV 426
+ KK + + + + + + + L VVE R+ GD Y+++
Sbjct: 362 MKKKKEEEYGFEMEGQN---GKFGIGICISRLNDLLVVVEARRRGGDGDCYKEM 412
>At4g14580 CBL-interacting protein kinase 4 (CIPK4)
Length = 426
Score = 343 bits (881), Expect = 8e-95
Identities = 178/401 (44%), Positives = 257/401 (63%), Gaps = 23/401 (5%)
Query: 15 VRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKL 74
V LGKYELG+ LG G+F KV +AR G+ A+KI++K K +D +I REI ++
Sbjct: 16 VLLGKYELGRRLGSGSFAKVHVARSISTGELVAIKIIDKQKTIDSGMEPRIIREIEAMRR 75
Query: 75 L-KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSY 133
L HPNV++++EV+A+K+KIY+V+EY GGELF K+ G+L E+ R+ FQQL +S+
Sbjct: 76 LHNHPNVLKIHEVMATKSKIYLVVEYAAGGELFTKLIRFGRLNESAARRYFQQLASALSF 135
Query: 134 CHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILA 193
CH G+ HRD+K +N+L+D +GN+K++DF LSALP+H +GLLHT CG+P Y APE++A
Sbjct: 136 CHRDGIAHRDVKPQNLLLDKQGNLKVSDFGLSALPEHRSNNGLLHTACGTPAYTAPEVIA 195
Query: 194 NRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIK 253
RGYDGAK+D WSCGV L+V+L GY+PFDD N+ +Y+KI K D + P W+S A++II
Sbjct: 196 QRGYDGAKADAWSCGVFLFVLLAGYVPFDDANIVAMYRKIHKRDYRFPSWISKPARSIIY 255
Query: 254 KILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSP 313
K+LDPNP+TR++++ + WF++ + + E D F++ EA
Sbjct: 256 KLLDPNPETRMSIEAVMGTVWFQKSLEISEFQSSVFELDRFLEKEA-------------- 301
Query: 314 KSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKV 373
KS I AF LI +SS LDLSG FE+ E+ RFT+ S + + EK I ++GF+V
Sbjct: 302 KSSNAITAFDLISLSSGLDLSGLFERRKRKEK--RFTARVSAERVVEKAGMIGEKLGFRV 359
Query: 374 HKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELR 414
KK + KV+ K +V EV E L V +++
Sbjct: 360 EKKE-ETKVVGLGKGR-----TAVVVEVVEFAEGLVVADVK 394
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.138 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,221,997
Number of Sequences: 26719
Number of extensions: 473608
Number of successful extensions: 4412
Number of sequences better than 10.0: 996
Number of HSP's better than 10.0 without gapping: 817
Number of HSP's successfully gapped in prelim test: 180
Number of HSP's that attempted gapping in prelim test: 1663
Number of HSP's gapped (non-prelim): 1124
length of query: 437
length of database: 11,318,596
effective HSP length: 102
effective length of query: 335
effective length of database: 8,593,258
effective search space: 2878741430
effective search space used: 2878741430
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC138579.3


