

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138465.9 - phase: 0
(521 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
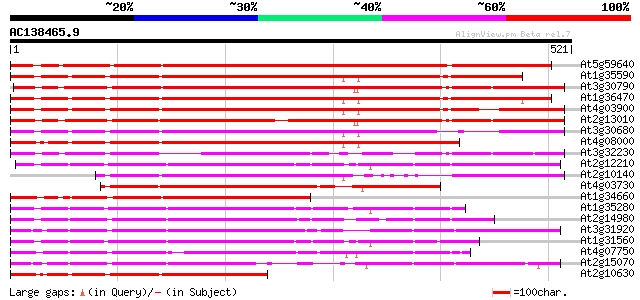
Score E
Sequences producing significant alignments: (bits) Value
At5g59640 serine/threonine-specific protein kinase - like 475 e-134
At1g35590 CACTA-element, putative 467 e-132
At3g30790 hypothetical protein 466 e-131
At1g36470 hypothetical protein 459 e-129
At4g03900 putative transposon protein 434 e-122
At2g13010 pseudogene 433 e-121
At3g30680 hypothetical protein 423 e-118
At4g08000 putative transposon protein 411 e-115
At3g32230 hypothetical protein 337 1e-92
At2g12210 putative TNP2-like transposon protein 325 4e-89
At2g10140 putative protein 305 3e-83
At4g03730 putative transposon protein 298 6e-81
At1g34660 hypothetical protein 292 3e-79
At1g35280 hypothetical protein 273 1e-73
At2g14980 pseudogene 273 2e-73
At3g31920 hypothetical protein 271 5e-73
At1g31560 putative protein 265 4e-71
At4g07750 putative transposon protein 249 3e-66
At2g15070 En/Spm-like transposon protein 239 3e-63
At2g10630 En/Spm transposon hypothetical protein 1 231 6e-61
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 475 bits (1223), Expect = e-134
Identities = 239/506 (47%), Positives = 322/506 (63%), Gaps = 20/506 (3%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVE-CKFCQEPRYKGRKHSRSSKGKPVP 59
M LGL ++ID C CM+F+ K+ E C FC++ RY+ ++ K +P
Sbjct: 127 MRSLGLPYQKIDVCQDNCMIFW------KETEKEEYCLFCKKDRYRP---TQKIGQKSIP 177
Query: 60 RKALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPN 119
+ +FYLPI RL+R+Y S TA+ M WH+++ + GE+ HPSDG AWKHF + HP+
Sbjct: 178 YRQMFYLPIADRLKRLYQSHNTAKHMRWHAEHLA---SDGEMGHPSDGEAWKHFHKVHPD 234
Query: 120 FALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGP 179
FA +PRNV LGLC+DGF P SG YS WP+I+TPYNLPP+MCM + +MFL ++PGP
Sbjct: 235 FASKPRNVYLGLCTDGFNPFGM-SGHNYSLWPVILTPYNLPPEMCMKQEFMFLTILVPGP 293
Query: 180 NNPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWG 238
N+PK +DI+LQPLI++LK LW NGV YDIS KQNF+++ LMWTI+ FP YGMLS W
Sbjct: 294 NHPKRSLDIFLQPLIEELKDLWVNGVEAYDISTKQNFLLKVVLMWTISDFPAYGMLSGWT 353
Query: 239 THGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPP 298
THGRLACP C + T AF L++G KT WF C R +LP +H +R NK F KG+ P
Sbjct: 354 THGRLACPYCSDQTGAFWLKNGRKTCWFDCHRCFLPVNHSYRGNKKDFKKGKVVEDSKPE 413
Query: 299 NLTSEQVWNKVKNKPKVQIVGGDASKPFGYGQKHNWTKRSIFWDLLYWKDNLLRHNLDVM 358
LT E+++N+V PK GG+ + GYG+ HNW K+SI W+L YWKD LRHNLDVM
Sbjct: 414 ILTGEELYNEVCCLPKTVDCGGNHGRLEGYGKTHNWHKQSILWELSYWKDLKLRHNLDVM 473
Query: 359 HIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYTLP 418
HIEKN DN T++NV+GKTKDN K+R D++ C RKDL L + P + L
Sbjct: 474 HIEKNVLDNFIKTLLNVQGKTKDNIKSRLDLQENCNRKDLHLTPEGKA----PIPKFRLK 529
Query: 419 IEETKDVYRWV-KELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTFSALE 477
+ + RW+ K++K SDGYSS+LA C D+ ++ GMKSHD H+ M+ L F+ L
Sbjct: 530 PDAKEIFLRWLEKDVKFSDGYSSSLANCVDLPGRKLTGMKSHDCHVLMQRLFPIAFAELM 589
Query: 478 PHVLNPLIEMSQYFKNSCSTTLREDD 503
++ + +++ E D
Sbjct: 590 DKSVHDALSRDRHYDGGNQEDTHEFD 615
>At1g35590 CACTA-element, putative
Length = 1180
Score = 467 bits (1201), Expect = e-132
Identities = 239/484 (49%), Positives = 314/484 (64%), Gaps = 25/484 (5%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
M LGL ID C CMLF+ +D +C+FC R+K + R +K VP
Sbjct: 242 MHNLGLPYHTIDVCKNNCMLFWK-----EDEKEDQCRFCGAQRWKPKDDRRRTK---VPY 293
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
++YLPI RL+RMY +TA M WH+ E ++ GE+ HPSD W++F + HP F
Sbjct: 294 SRMWYLPIADRLKRMYQCHKTAAAMRWHA---EHQSKEGEMNHPSDAAEWRYFQELHPRF 350
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A EPRNV LGLC+DGF P G + +S WP+I+TPYNLPP MCM+ Y+FL + GPN
Sbjct: 351 AEEPRNVYLGLCTDGFNPF--GMSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNYGPN 408
Query: 181 NPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+P+ +D++LQPLI++LK LW GV Y++S QNF ++A L+WTI+ FP Y MLS W T
Sbjct: 409 HPRASLDVFLQPLIEELKELWCTGVDAYEVSLSQNFNLKAVLLWTISDFPAYNMLSGWTT 468
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKG-ETENRGPPP 298
HG+L+CP+CME T +F L G KT WF C RR+LP HP RRNK F+KG + + PP
Sbjct: 469 HGKLSCPVCMESTKSFYLPIGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEYPPE 528
Query: 299 NLTSEQVWNK---VKNKPKVQIVGGDA--SKPFGYGQKHNWTKRSIFWDLLYWKDNLLRH 353
+LT EQV+ + N PK + VGG+ K GYG++HNW K SI W+L YWKD LRH
Sbjct: 529 SLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRH 588
Query: 354 NLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCA 413
N+DVMH EKNF DNI NT++ VKGK+KDN +R DIE YC R L + S+GK P
Sbjct: 589 NIDVMHTEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHI--DSTGK--APFP 644
Query: 414 NYTLPIEETKDVYRWVK-ELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTT 472
YTL + + +++ VK +++ DGYSS+LA C D+ENG+ GMKSHD HIFME L+
Sbjct: 645 PYTLTEQAKQSLFQCVKHDVRFPDGYSSDLASCVDLENGKFSGMKSHDCHIFMERLLPFI 704
Query: 473 FSAL 476
F+ L
Sbjct: 705 FAEL 708
>At3g30790 hypothetical protein
Length = 953
Score = 466 bits (1200), Expect = e-131
Identities = 244/521 (46%), Positives = 332/521 (62%), Gaps = 28/521 (5%)
Query: 4 LGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPRKAL 63
LGL + ID C+ CM+F+ + D +L EC+FC +PRY+ +S VP K +
Sbjct: 146 LGLPSEMIDVCIDYCMIFWGD-----DVNLQECRFCGKPRYQ-----TTSGRTRVPYKRM 195
Query: 64 FYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNFALE 123
+YLPI RL+R+Y S +TA KM WH+Q+ GE++HPSD AWKHF +P+FA E
Sbjct: 196 WYLPITDRLKRLYQSERTAAKMRWHAQH---TTADGEIKHPSDTKAWKHFQTVYPDFASE 252
Query: 124 PRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPNNPK 183
RNV LGLC+DGF+P + SG+ YS W II+TPYNLPPDM M ++FL+ ++PGP +PK
Sbjct: 253 CRNVYLGLCTDGFSPFVL-SGRQYSLWHIILTPYNLPPDMYMQSEFLFLSILVPGPKHPK 311
Query: 184 VVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGTHGR 242
+D++LQPLI +LK+LW GVHT+D S KQNF MRA LMWTI+ FP YGMLS W THGR
Sbjct: 312 RALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAVLMWTISDFPAYGMLSGWTTHGR 371
Query: 243 LACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPNLTS 302
L+CP CM TDAF+L+ G K WF RR+LP HP+RRNK F K + PP ++
Sbjct: 372 LSCPYCMGRTDAFQLKKGRKPCWFDYHRRFLPLHHPYRRNKKLFRKNKIVRLPPPSYVSG 431
Query: 303 EQVWNKVKNKPKVQIV--GGD----ASKPFGYGQKHNWTKRSIFWDLLYWKDNLLRHNLD 356
++ ++ + GG+ A+ P YG HNW K+SIFW L YWKD LRH LD
Sbjct: 432 TDLFKQIDYYGAQETCKRGGNWLTTANMPDEYGSTHNWHKQSIFWQLPYWKDLFLRHCLD 491
Query: 357 VMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYT 416
VMHIEKNFFDNI NT++NV+GKTKD+ K+R D+E C R +L + GK P + +
Sbjct: 492 VMHIEKNFFDNIMNTLLNVQGKTKDSLKSRLDLEEICSRPELHVTR--DGKL--PVSKFR 547
Query: 417 LPIEETKDVYRW-VKELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTFSA 475
L E K ++ W V E+K DGY S +RC + + + GMKSHD H+FM+ L+
Sbjct: 548 LSNEAKKALFEWVVAEVKFPDGYVSKFSRCVE-QGQKFSGMKSHDCHVFMQRLLPFVLVE 606
Query: 476 LEP-HVLNPLIEMSQYFKNSCSTTLREDDLIKMENEIPMIL 515
L P ++ + + +FK+ C+ TL + +++ IP++L
Sbjct: 607 LLPTNIHEAIAGIGVFFKDLCTRTLTTYTIEQLDKNIPVLL 647
>At1g36470 hypothetical protein
Length = 753
Score = 459 bits (1182), Expect = e-129
Identities = 241/520 (46%), Positives = 322/520 (61%), Gaps = 34/520 (6%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
M LGL ID C CML + +D +C+FC R+K + R +K VP
Sbjct: 74 MRNLGLPYHTIDVCQNNCMLSWK-----EDEKEDQCRFCGAKRWKPKDDRRRTK---VPY 125
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
++YLPI RL+RMY S +TA M WH+ E ++ GE+ H SD W++F HP F
Sbjct: 126 SRMWYLPIGDRLKRMYQSHKTAAAMRWHA---EHQSKEGEMNHLSDAAEWRYFQGLHPEF 182
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A EPRNV LGLC+DGF P G + +S WP+I+TPYNLPP MCM+ Y+FL + GPN
Sbjct: 183 AEEPRNVYLGLCTDGFNPF--GMSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPN 240
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+P+ +D++LQPLI++LK W+ GV YD+S QNF ++A L+WTI+ FP Y ML W T
Sbjct: 241 HPRASLDVFLQPLIEELKEFWSTGVDAYDVSLSQNFNLKAVLLWTISDFPAYNMLLGWTT 300
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKG-ETENRGPPP 298
HG+L+CP+CME T +F L +G KT WF C RR+LP HP RRN+ F+KG + + PP
Sbjct: 301 HGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNRKDFLKGRDASSEYPPE 360
Query: 299 NLTSEQVWNK---VKNKPKVQIVGGDA--SKPFGYGQKHNWTKRSIFWDLLYWKDNLLRH 353
+LT EQV+ + N PK + VGG+ K GYG++HNW K SI W+L YWKD LRH
Sbjct: 361 SLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRH 420
Query: 354 NLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCA 413
N+DVMH EKNF DNI NT+M VKGK+KDN +R DIE +C R L + SSGK P
Sbjct: 421 NIDVMHTEKNFLDNIMNTLMRVKGKSKDNIMSRLDIEKFCSRPGLHI--DSSGK--APFP 476
Query: 414 NYTLPIEETKDVYRWVK-ELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTT 472
YTL E + + + VK +++ DGYSS+LA C D++NG+ GMKSHD H+FME L+
Sbjct: 477 AYTLTEEAKQSLLQCVKYDIRFPDGYSSDLASCVDLDNGKFSGMKSHDCHVFMERLLPFI 536
Query: 473 FS---------ALEPHVLNPLIEMSQYFKNSCSTTLREDD 503
F+ AL ++L +S C+ T + +
Sbjct: 537 FAELLDRNVHLALSGNILEHYFSISYLGSRDCARTFEQGE 576
>At4g03900 putative transposon protein
Length = 817
Score = 434 bits (1115), Expect = e-122
Identities = 235/524 (44%), Positives = 319/524 (60%), Gaps = 44/524 (8%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
M LGL ID CMLF+ +D +C+FC R+K + R +K VP
Sbjct: 242 MRNLGLPYHTIDVYKNNCMLFWK-----EDEKEDQCRFCGAQRWKPKDDRRRTK---VPY 293
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
++YLPI RL+RMY S +TA M WH+ E ++ GE+ HPSD W++F + HP F
Sbjct: 294 SRMWYLPIADRLKRMYQSHKTAAAMRWHA---EHQSKEGEMNHPSDAAEWRYFQELHPRF 350
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A EP NV LGL +DGF P G + +S WP+I+TPYNLPP MCM+ Y+FL + GPN
Sbjct: 351 AEEPHNVYLGLYTDGFNPF--GMSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPN 408
Query: 181 NPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+P+ +D++LQPLI++LK LW GV YD+S QNF ++A L+WTI+ FP Y MLS W T
Sbjct: 409 HPRASLDVFLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTT 468
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKG-ETENRGPPP 298
HG+L+CP+CME T +F L +G KT WF C RR+LP HP RRNK F+KG + + PP
Sbjct: 469 HGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEYPPE 528
Query: 299 NLTSEQVWNK---VKNKPKVQIVGGDA--SKPFGYGQKHNWTKRSIFWDLLYWKDNLLRH 353
+LT EQV+ + N PK + VGG+ K GYG++HNW K SI W+L YWKD LRH
Sbjct: 529 SLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRH 588
Query: 354 NLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCA 413
N+DVMH EKNF DNI NT++ VKGK+KDN +R DIE YC R L + S+GK P
Sbjct: 589 NIDVMHTEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPGLHI--DSTGK--APFP 644
Query: 414 NYTLPIEETKDVYRWVK-ELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTT 472
Y L E + +++ VK +++ D GMKSHD H+FME L+
Sbjct: 645 PYKLTEEAKQSLFQCVKHDVRFPD------------------GMKSHDCHVFMERLLPFI 686
Query: 473 FS-ALEPHVLNPLIEMSQYFKNSCSTTLREDDLIKMENEIPMIL 515
F+ L+ +V L + +F++ CS TL+ + ++ I +I+
Sbjct: 687 FAELLDRNVHLALSGIRAFFRDLCSRTLQTSRVQILKQNIVLII 730
>At2g13010 pseudogene
Length = 1040
Score = 433 bits (1114), Expect = e-121
Identities = 233/525 (44%), Positives = 320/525 (60%), Gaps = 41/525 (7%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKP-VP 59
+S GL + ID C+ CM+F+ + D +L EC+FC +PRY+ ++ G+ VP
Sbjct: 393 VSSHGLPSEMIDVCIDYCMIFWGD-----DVNLQECRFCGKPRYQ------TTGGRTRVP 441
Query: 60 RKALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPN 119
K ++YLPI RL+R+Y S +TA KM WH+Q+ GE+ HPSD AWKHF +P+
Sbjct: 442 YKRMWYLPITDRLKRLYQSERTAAKMRWHAQH---TTADGEITHPSDAKAWKHFQTVYPD 498
Query: 120 FALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGP 179
FA E RNV LGLC+DGF+P G+ YS WP+I+TPY LPPDMCM +MFL+ ++PGP
Sbjct: 499 FANECRNVYLGLCTDGFSP-FGLCGRQYSLWPVILTPYKLPPDMCMQSEFMFLSILVPGP 557
Query: 180 NNPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWG 238
+PK +D++LQPLI +LK+LW GVHT+D S KQNF MRA LMWTI+ FP Y MLS W
Sbjct: 558 KHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAMLMWTISDFPAYRMLSGWT 617
Query: 239 THGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPP 298
HGRL P K WF C RR+LP HP+RRNK F K + PP
Sbjct: 618 MHGRLYVPT-----------KDRKPCWFNCHRRFLPLHHPYRRNKKLFRKNKIVRLLPPS 666
Query: 299 NLTSEQVWNKVKNKPKVQIV--GGD----ASKPFGYGQKHNWTKRSIFWDLLYWKDNLLR 352
++ ++ ++ + GG+ A+ P YG HNW K+SIFW L YWKD LLR
Sbjct: 667 YVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDEYGSTHNWYKQSIFWQLPYWKDLLLR 726
Query: 353 HNLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPC 412
HNL VMHIEKNFFDNI NT++N++GKTKD K+R D+ C + +L + GK P
Sbjct: 727 HNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRLDLAEICSKPELHVTR--DGKL--PV 782
Query: 413 ANYTLPIEETKDVYRW-VKELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLIST 471
+ + L E K ++ W V E+K DGY S +RC + + + GMKSHD H+FM+ L+
Sbjct: 783 SKFRLSNEAKKALFEWVVAEVKFPDGYVSKFSRCVE-QGQKFSGMKSHDCHVFMQRLLPF 841
Query: 472 TFSALEP-HVLNPLIEMSQYFKNSCSTTLREDDLIKMENEIPMIL 515
L P ++ + + +FK+ C+ TL D + +++ IP++L
Sbjct: 842 VLVELLPTNIHEAIAGIGVFFKDLCTRTLTTDTIEQLDKNIPVLL 886
>At3g30680 hypothetical protein
Length = 1116
Score = 423 bits (1088), Expect = e-118
Identities = 228/523 (43%), Positives = 305/523 (57%), Gaps = 72/523 (13%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
M LGL ID C CMLF+ +D +C+FC R+K + R +K VP
Sbjct: 242 MRNLGLPYHTIDVCKNNCMLFWK-----EDEKEDQCRFCGAQRWKPKDDRRRTK---VPY 293
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
++YLPI +L+RMY S +TA M WH+ E ++ GE+ HPSD W++F + HP F
Sbjct: 294 SRMWYLPIADQLKRMYQSHKTAAAMRWHA---EHQSKEGEMNHPSDAAEWRYFQELHPRF 350
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A EPRNV LGLC+DGF P + +S WP+I+TPYNLPP MCM+ Y+FL + GPN
Sbjct: 351 AEEPRNVYLGLCTDGFNPF--SMSRNHSLWPVILTPYNLPPRMCMNTEYLFLTILNSGPN 408
Query: 181 NPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+P+ +D++LQPLI++LK LW GV YD+S QNF ++A L+WTI+ FP Y MLS W T
Sbjct: 409 HPRASLDVFLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTT 468
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKG-ETENRGPPP 298
HG+L+CP+CME T +F L +G KT WF C RR+LP HP RRNK F+KG + + PP
Sbjct: 469 HGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEYPPE 528
Query: 299 NLTSEQVWNK---VKNKPKVQIVGGDA--SKPFGYGQKHNWTKRSIFWDLLYWKDNLLRH 353
+LT EQV+ + N PK + VGG+ K GYG++HNW K SI W+L YWKD LRH
Sbjct: 529 SLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRH 588
Query: 354 NLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCA 413
N+DVMH EKNF DNI NT++ VKGK+KDN +R DIE YC R
Sbjct: 589 NIDVMHTEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYCSRPG---------------- 632
Query: 414 NYTLPIEETKDVYRWVKELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTF 473
L I+ T GMKSHD H+FME L+ F
Sbjct: 633 ---LHIDST--------------------------------GMKSHDCHVFMERLLPFIF 657
Query: 474 S-ALEPHVLNPLIEMSQYFKNSCSTTLREDDLIKMENEIPMIL 515
+ L+ +V L + +F++ CS TL+ + ++ I +I+
Sbjct: 658 AELLDRNVHLALSGIGAFFRDLCSRTLQTSRVQILKQNIVLII 700
>At4g08000 putative transposon protein
Length = 609
Score = 411 bits (1056), Expect = e-115
Identities = 206/424 (48%), Positives = 275/424 (64%), Gaps = 20/424 (4%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
M LGL ID C CMLF+ + ++ H C+FC R+K + R +K VP
Sbjct: 96 MRNLGLPYHTIDVCSNNCMLFWKED--EQEDH---CRFCGAQRWKPKDDRRRTK---VPY 147
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
++YLPI RL+RMY S +TA M WH+ E ++ GE+ HPSD W++F + HP F
Sbjct: 148 SRMWYLPIGDRLKRMYQSHKTAAAMRWHA---EHQSKEGEMNHPSDAAEWRYFQELHPRF 204
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A EPRNV LGLC+DGF P G + +S WP+I+TPYNLPP MCM+ Y+FL + PN
Sbjct: 205 AEEPRNVYLGLCTDGFNPF--GMSRNHSLWPVILTPYNLPPSMCMNTKYLFLTILNVEPN 262
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+P+ +D++LQPLI++LK LW+ GV YD+S QNF ++A L+WTI+ FP Y MLS W T
Sbjct: 263 HPRASLDVFLQPLIEELKELWSTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWIT 322
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKG-ETENRGPPP 298
HG+ +CPICME T +F L +G KT WF C RR+LP HP RRNK F+KG ++ + PP
Sbjct: 323 HGKFSCPICMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDSSSEYPPK 382
Query: 299 NLTSEQVWNK---VKNKPKVQIVGGDA--SKPFGYGQKHNWTKRSIFWDLLYWKDNLLRH 353
+LT EQV+ + N K + VGG+ K GYG++HNW K SI W+L YW D LRH
Sbjct: 383 SLTGEQVYYERLASVNPTKTKDVGGNGHEKKMPGYGKEHNWHKESILWELSYWNDLNLRH 442
Query: 354 NLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCA 413
N+DVMH EKNF D+I NT+M+VK K+KDN +R DIE++C R DL + + G F +
Sbjct: 443 NIDVMHTEKNFLDDIMNTLMSVKDKSKDNIMSRLDIEIFCSRSDLHIDSQGIGAFFRDLC 502
Query: 414 NYTL 417
+ TL
Sbjct: 503 SRTL 506
>At3g32230 hypothetical protein
Length = 1169
Score = 337 bits (863), Expect = 1e-92
Identities = 205/517 (39%), Positives = 274/517 (52%), Gaps = 100/517 (19%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+S LGL + ID C+ CM+F+ + D +L EC+FC +PRY+ + + VP
Sbjct: 330 VSSLGLPSEMIDVCIDYCMIFWKD-----DENLQECRFCGKPRYQP-----TGRRTRVPY 379
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K ++YLPI RL+R+Y S +TA KM WH+ E +GE+ HPSD AWKHF +P+F
Sbjct: 380 KRMWYLPITDRLKRLYQSERTAAKMRWHA---EHSTVNGEVTHPSDAKAWKHFQTVYPDF 436
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A E RNV LGLC+DGF+P GP
Sbjct: 437 ASECRNVYLGLCTDGFSPF--------------------------------------GPK 458
Query: 181 NPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+PK +D++LQPLI +LK LW GV T+D S KQNF MRA LMWTI+ FP YGMLS W T
Sbjct: 459 HPKRALDVFLQPLIHELKNLWYEGVQTFDYSSKQNFNMRAMLMWTISDFPAYGMLSGWTT 518
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPN 299
HGRL+CP CM+ TDAF+L+HG K+ +L + F N + + ET RG
Sbjct: 519 HGRLSCPYCMDRTDAFQLKHGRKSFVRLPPPVYLSGNDLF-ENIDYYGSQETCKRGGN-- 575
Query: 300 LTSEQVWNKVKNKPKVQIVGGDASKPFGYGQKHNWTKRSIFWDLLYWKDNLLRHNLDVMH 359
W+ N P GYG HNW K+SIFW L YWKD LLRHNLD
Sbjct: 576 ------WHTPANMPD------------GYGSAHNWHKQSIFWQLPYWKDLLLRHNLD--- 614
Query: 360 IEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYTLPI 419
GKTKD+ K+R D+ C R DL + GK P + L
Sbjct: 615 -----------------GKTKDSMKSRLDLAEICNRPDLHITR--DGKL--PVPKFRLTT 653
Query: 420 EETKDVYRWVK-ELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTFSALEP 478
E K ++ WVK E+K DGY S +RC + + + GMKSHD H+F++ L+ F L P
Sbjct: 654 EGKKALFEWVKAEVKFPDGYLSKFSRCVE-QGQKFSGMKSHDCHVFIQRLLPFVFLELLP 712
Query: 479 HVLNPLIEMSQYFKNSCSTTLREDDLIKMENEIPMIL 515
++ IE +F++ C+ TL D + +++ IPM+L
Sbjct: 713 ANVHDAIE-GAFFRDLCTRTLTTDGIQQLDENIPMLL 748
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 325 bits (833), Expect = 4e-89
Identities = 196/539 (36%), Positives = 289/539 (53%), Gaps = 51/539 (9%)
Query: 6 LGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPRKALFY 65
+G ++I CV C LF + +L C C R+K + +K K VP+K L Y
Sbjct: 199 MGYEKIHACVNDCCLFRK-----RYKNLENCPKCNASRWKSNMQTGEAK-KGVPQKVLRY 252
Query: 66 LPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNFALEPR 125
PIIPR +RM+ S + A+ + WH Y ++ G+LRHP D + W + K+P FA+E R
Sbjct: 253 FPIIPRFKRMFRSEEMAKDLRWH---YSNKSTDGKLRHPVDSVTWDKMNDKYPEFAIEER 309
Query: 126 NVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPNNPKVV 185
N+RLGL +DGF P + + YSCWP+++ YNLPPD+CM K + L+ +IPGP P
Sbjct: 310 NLRLGLSTDGFNPFNMKNTR-YSCWPVLLVNYNLPPDLCMKKENIMLSLLIPGPQQPGNS 368
Query: 186 IDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGTHGRLA 244
ID+YL+PLI+DL LW G TYD K F ++A L+WTI+ FP YG L+ G++
Sbjct: 369 IDVYLEPLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTISDFPAYGNLAGCKVKGKMG 428
Query: 245 CPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPNLTSEQ 304
CP+C + TD+ L++ K F C R+ L H + RNK A+ G+ E+ LT +
Sbjct: 429 CPLCGKGTDSMWLKYSRKHV-FMCHRKGLAPTHSY-RNKKAWFDGKIEHGRKARILTGRE 486
Query: 305 VWNKVKNKPKVQIVGGDASKPFGYGQKHN------------------------------- 333
V +KN + G+A K G +K N
Sbjct: 487 VSQTLKN---FKNDFGNA-KESGRKRKRNDCIESVTDSDDESSESEEDEELEVDEDELSR 542
Query: 334 WTKRSIFWDLLYWKDNLLRHNLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYC 393
W KRSIF+ L YW+D +RHNLDVMH+E+N +I +T+++ GK+KD ARKD++
Sbjct: 543 WKKRSIFFKLSYWEDLPVRHNLDVMHVERNVGASIVSTLLHC-GKSKDGLNARKDLQDLG 601
Query: 394 RRKDLLLVEKSSGKFLKPCANYTLPIEETKDVYRWVKELKMSDGYSSNLARCADVENGRM 453
RKDL + +L P A ++L E K + + E K DGY SN++R +E+ ++
Sbjct: 602 VRKDLHPNAQGKRTYL-PAAPWSLSKSEKKTFCKRLFEFKGPDGYCSNISRGVSLEDCKI 660
Query: 454 HGMKSHDTHIFMECLISTTFSALEPH-VLNPLIEMSQYFKNSCSTTLREDDLIKMENEI 511
G+KSHD H+ M+ L+ L P +I + +F + C + + + ME EI
Sbjct: 661 MGLKSHDYHVLMQQLLPVALMGLLPKGPRTAIIRLCSFFNHLCQRVIDIEVISVMEAEI 719
>At2g10140 putative protein
Length = 663
Score = 305 bits (782), Expect = 3e-83
Identities = 178/443 (40%), Positives = 247/443 (55%), Gaps = 76/443 (17%)
Query: 80 QTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNFALEPRNVRLGLCSDGFTPH 139
+TA M WH+ E ++ GE+ HPSD W++F + H FA EPRNV LGLC DGF P
Sbjct: 244 KTAAAMRWHA---EHQSKEGEMNHPSDAAEWRYFQELHTWFAKEPRNVYLGLCIDGFNPF 300
Query: 140 IQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPNNPKVVIDIYLQPLIDDLKR 199
G + +S WP+I+T YNLPP MCM+ Y+FL + GPN+P+ +D++LQPLI++LK
Sbjct: 301 --GISRNHSLWPVILTSYNLPPGMCMNTKYLFLTILNSGPNHPRASLDVFLQPLIEELKE 358
Query: 200 LW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGTHGRLACPICMEDTDAFRLE 258
LW GV TYD+S QNF ++A L+W I+ FP Y MLS W THG+L+CP+CME T +F L
Sbjct: 359 LWCTGVDTYDVSLSQNFNLKAVLLWAISDFPAYSMLSGWTTHGKLSCPVCMESTKSFYLP 418
Query: 259 HGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRG-PPPNLTSEQVWNK---VKNKPK 314
+G KT WF C RR+LP HP RRNK F+KG +R PP +LT EQV+ + N PK
Sbjct: 419 NGRKTCWFDCHRRFLPHSHPSRRNKKDFLKGRDASREYPPGSLTGEQVYYERLASVNPPK 478
Query: 315 VQIVGGDASKPFGYGQKHNWTKRSIFWDLLYWKDNLLRHNLDVMHIEKNFFDNIFNTVMN 374
+ VG G H R Y K+ HN D ++IF
Sbjct: 479 TKDVG---------GNGHEKKMRG------YGKE----HNWDK--------ESIFT---- 507
Query: 375 VKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYTLPIEETKDVYRWVK-ELK 433
GKT P YTL E + ++++VK +++
Sbjct: 508 --GKT-------------------------------PFPAYTLIEEGKQSLFQFVKHDVR 534
Query: 434 MSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTFS-ALEPHVLNPLIEMSQYFK 492
DG+SS+LA C D+ENG+ GMKSHD H+FME L+ F+ L+ +V L + +F+
Sbjct: 535 FPDGHSSDLASCVDLENGKFSGMKSHDCHVFMERLLPFIFAELLDRNVHLALSGIGAFFR 594
Query: 493 NSCSTTLREDDLIKMENEIPMIL 515
+ CS TL+ + ++ I +I+
Sbjct: 595 DLCSRTLQISRVQILKQNIVLII 617
>At4g03730 putative transposon protein
Length = 696
Score = 298 bits (762), Expect = 6e-81
Identities = 151/322 (46%), Positives = 201/322 (61%), Gaps = 26/322 (8%)
Query: 85 MTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNFALEPRNVRLGLCSDGFTPHIQGSG 144
M WH+ E + GE+ +PSDG AWK+F++ +PNFA E RN+ LGL +DGF + +G
Sbjct: 3 MRWHA---EHMSPEGEMHNPSDGAAWKNFNEVYPNFAAESRNIYLGLSTDGFNL-VSMNG 58
Query: 145 KPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPNNPKVVIDIYLQPLIDDLKRLW-NG 203
+ +S W +IVTPYNLP MC+ + + FL+ ++PGP +PK +DIYLQPLI +L LW +G
Sbjct: 59 EAHSVWHVIVTPYNLPSGMCIKREFFFLSVLVPGPKHPKKNLDIYLQPLIKELVSLWIDG 118
Query: 204 VHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGTHGRLACPICMEDTDAFRLEHGGKT 263
YD S+K+ F MR AL W I+ F TYGMLS W THG L+CP C+++T +F L + K
Sbjct: 119 EEAYDKSKKKKFTMRVALTWAISDFSTYGMLSVWMTHGLLSCPYCLDNTISFWLPNERKH 178
Query: 264 TWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPNLTSEQVWNKVKNKPKVQIVGGDAS 323
+WF C +LP H + +N AF +G E R PP GG+
Sbjct: 179 SWFDCHGMFLPLGHMYMQNVQAF-RGGKEVRDDPPFTVD---------------CGGNEH 222
Query: 324 KPF-----GYGQKHNWTKRSIFWDLLYWKDNLLRHNLDVMHIEKNFFDNIFNTVMNVKGK 378
+ F GYG HNW KRSIFW+L YW++ +LRHNLD MHI+KNFFDN+ NTV+ V K
Sbjct: 223 EKFAQTIDGYGIHHNWVKRSIFWELPYWENLILRHNLDFMHIKKNFFDNLINTVLKVLEK 282
Query: 379 TKDNEKARKDIELYCRRKDLLL 400
TKD K+R D+ C R L L
Sbjct: 283 TKDKIKSRMDLATLCNRSGLHL 304
>At1g34660 hypothetical protein
Length = 501
Score = 292 bits (748), Expect = 3e-79
Identities = 140/280 (50%), Positives = 194/280 (69%), Gaps = 16/280 (5%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+S LGL + ID CV CM+F+ + +L C FCQ+PR++ S K + VP
Sbjct: 212 VSSLGLPYQMIDVCVDNCMIFWRDTV-----NLDACTFCQKPRFQ----VTSGKSR-VPF 261
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K ++YLPI RL+R+Y S +TA M WH+++ ++ G + HPSD AWKHF+ + +F
Sbjct: 262 KRMWYLPITDRLKRLYQSERTAGPMRWHAEH----SSDGNISHPSDAEAWKHFNSMYKDF 317
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A E RNV LGLC+DGF P + SG+ YS WP+I+TPYNLPP +CM + ++FL+ ++PGP
Sbjct: 318 ANEHRNVYLGLCTDGFNPFGK-SGRKYSLWPVILTPYNLPPSLCMKREFLFLSILVPGPE 376
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+P+ +D++LQPLI +L+ LW+ GV YD+S K NFVMRA LMWTI+ FP YGMLS W T
Sbjct: 377 HPRRSLDVFLQPLIHELQLLWSEGVVAYDVSLKNNFVMRAVLMWTISDFPAYGMLSGWTT 436
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPF 279
HGRL+CP C + DAF+L+HG K+ WF C RR+LP +H +
Sbjct: 437 HGRLSCPYCQDTIDAFQLKHGKKSCWFDCHRRFLPKNHSY 476
>At1g35280 hypothetical protein
Length = 557
Score = 273 bits (699), Expect = 1e-73
Identities = 166/455 (36%), Positives = 239/455 (52%), Gaps = 50/455 (10%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+ +G ++I CV C LF + +L C C R+K + +K K VP+
Sbjct: 19 LKSFDMGYEKIHACVNDCCLFRK-----RYKNLENCPKCNASRWKSNMQTGEAK-KGVPQ 72
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K L Y PIIPR +RM+ S + A+ + WH Y ++ G+LRHP D + W + K+P F
Sbjct: 73 KVLRYFPIIPRFKRMFRSEEMAKDLRWH---YSNKSIDGKLRHPVDSVTWDKMNDKYPEF 129
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A E RN+RLGL +DGF P + + YSCWP+++ YNLP D+CM K + L+ +IPGP
Sbjct: 130 AAEERNLRLGLSTDGFNPFNMKNTR-YSCWPVLLVNYNLPLDLCMKKENIMLSLLIPGPQ 188
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
P ID+YL+PLI+DL LW G TYD K F ++A L+WTI+ F YG L+
Sbjct: 189 QPGNSIDVYLEPLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTISDFSAYGNLAGCKV 248
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPN 299
G++ CP+C + TD+ L++ K F C R+ L H +R NK A+ G+ E+
Sbjct: 249 KGKMGCPLCGKGTDSMWLKYSRKHV-FMCHRKGLAPTHSYR-NKKAWFDGKIEHGRKARI 306
Query: 300 LTSEQVWNKVKNKPKVQIVGGDASKPFGYGQKHN-------------------------- 333
LT +V +KN +K G +K N
Sbjct: 307 LTGREVSQTLKNFKN----DFRNAKESGRKRKRNDCIESVTDSDDESSESEEDEELEVDK 362
Query: 334 -----WTKRSIFWDLLYWKDNLLRHNLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKD 388
W KRSIF+ L YW+D +RHNLDVMH+E+N +I +T+++ GK+KD ARKD
Sbjct: 363 DELSRWKKRSIFFKLSYWEDLPVRHNLDVMHVERNVGASIVSTLLHC-GKSKDGLNARKD 421
Query: 389 IELYCRRKDLLLVEKSSGKFLKPCANYTLPIEETK 423
++ RKDL + +L P A ++L E K
Sbjct: 422 LQDLGVRKDLHPNAQGKRTYL-PAAPWSLSKSERK 455
>At2g14980 pseudogene
Length = 665
Score = 273 bits (698), Expect = 2e-73
Identities = 167/451 (37%), Positives = 245/451 (54%), Gaps = 31/451 (6%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+ +G ++I CV C LF K L C C R+K H+ K K VP+
Sbjct: 87 LKSFDMGYQKIHACVNDCCLFRK-----KYKKLQNCLKCNASRWKTNMHTGEVK-KGVPQ 140
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K L Y I+PRL+RM+ S + A + WH N ++ G+L HP D + W + K+P F
Sbjct: 141 KVLRYFSIVPRLKRMFRSEEMARDLRWHFHN---KSTDGKLCHPVDSVTWDQMNAKYPLF 197
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A E RN+RLGL ++GF P + + YSCWP+++ YNLPPD+CM K + L +IPG
Sbjct: 198 ASEERNLRLGLSTNGFNPFNMKNTR-YSCWPVLLVNYNLPPDLCMKKENIMLTLLIPGSQ 256
Query: 181 NPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
P ID+YL+PLIDDL LW NG TYD K +F ++A L+WTI+ FP YG L+
Sbjct: 257 QPGNSIDVYLEPLIDDLCHLWDNGEWTYDAFSKSSFTLKAMLLWTISDFPAYGNLAGCKV 316
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPN 299
G++AC +C + T++ L+ K + G R LP H F RNK ++ G+ E+R
Sbjct: 317 KGKMACLLCGKHTESMWLKFSRKHVYMG-HRMGLPPSHRF-RNKKSWFDGKAEHRRKSRI 374
Query: 300 LTSEQVWNKVKNKPKVQIVGGDASKPFGYGQKHNWTKRSIFWDLLYWKDNLLRHNLDVMH 359
L+ ++ + +KN FG ++ ++ DL +RHNLDVMH
Sbjct: 375 LSGLEISHNLKN----------FQNNFGNFKQSTRKRKQPESDL------PVRHNLDVMH 418
Query: 360 IEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYTLPI 419
+E+N +I +T+M+ GK+KD ARKD+E+ RKDL K +L P A ++L
Sbjct: 419 VERNVVASIVSTLMHC-GKSKDGVNARKDLEVLGIRKDLHPQTKGKRTYL-PAALWSLSK 476
Query: 420 EETKDVYRWVKELKMSDGYSSNLARCADVEN 450
E K + + + K DGY S++AR +E+
Sbjct: 477 SEKKLFCKRLFDFKGPDGYCSDIARGVSLED 507
>At3g31920 hypothetical protein
Length = 725
Score = 271 bits (694), Expect = 5e-73
Identities = 172/514 (33%), Positives = 254/514 (48%), Gaps = 42/514 (8%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+ G G I C C+L Y E+ ++D C C R++ KH+ K K +P
Sbjct: 202 LKMFGFGYDIIHACQNDCIL-YRKEYELRD----TCPRCNASRWERDKHTGEEK-KGIPA 255
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K L Y PI R +RM+ S + AE WH N ++ G +RH D LAW + K P F
Sbjct: 256 KVLGYFPIKDRFKRMFRSKKMAEDQRWHFSN---ASDDGTMRHHVDSLAWAQVNDKWPEF 312
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A EPRN+RLGL +DG P + K YS WPI++ YN+ P MCM + L +I GP
Sbjct: 313 AAEPRNLRLGLSADGMNPFSIQNTK-YSTWPILLVNYNMAPTMCMKAENIMLTLLIHGPT 371
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
P ID+YL PLIDDLK LW+ G+H YD +K++F +RA +W+I+ +P G L+
Sbjct: 372 APSNNIDVYLAPLIDDLKDLWSEGIHVYDSFKKESFTLRAMFLWSISDYPALGTLAGCKV 431
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPN 299
G+ AC +C +DT L+ K + RR P HP+RR + F T G
Sbjct: 432 KGKQACNVCGKDTPFRWLKFSRKHVYLRNRRRLRP-GHPYRRRRGWF--DNTVEEGTTSR 488
Query: 300 L-TSEQVWNKVKNKPKVQIVGGDASKPFGYGQKHNWTKRSIFWDLLYWKDNLLRHNLDVM 358
+ T E+++ +K + I DL D +RHN+DVM
Sbjct: 489 IQTGEEIFEALK------------------------ALKMILGDLWRETDMPVRHNIDVM 524
Query: 359 HIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYTLP 418
H+EKN D I + +M K+KD ARKD+E R +L + +L P A Y L
Sbjct: 525 HVEKNVSDAIVSMLMQ-SVKSKDGLNARKDLEDMGTRSNLHPQLRVKRTYLPPAA-YWLS 582
Query: 419 IEETKDVYRWVKELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTFSALEP 478
+E + + + + DGY +N++ C ++ + G+KSHD H+ ++ L+ L P
Sbjct: 583 KDEKRRFCKRLARFRGPDGYCANISNCVSMDPPVIGGLKSHDHHVLLQNLLPVALRGLLP 642
Query: 479 HVLNPLI-EMSQYFKNSCSTTLREDDLIKMENEI 511
V I + YF C + + L+ +E+E+
Sbjct: 643 RVPRVAISRLCHYFNRLCQRVIDPEKLMSLESEL 676
>At1g31560 putative protein
Length = 1431
Score = 265 bits (678), Expect = 4e-71
Identities = 166/469 (35%), Positives = 243/469 (51%), Gaps = 52/469 (11%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+ +G ++I CV C LF + +L C C R+K + +K K VP+
Sbjct: 125 LKSFDMGYEKIHACVNDCCLFRK-----RYKNLENCPKCNASRWKNYMQTGEAK-KGVPQ 178
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K L Y IIPR ++M+ S + A+ + WH Y ++ G+ RHP D + W K+P F
Sbjct: 179 KVLRYFLIIPRFKKMFRSKEMAKDLRWH---YSNKSTDGKHRHPVDSVTWDKMIDKYPEF 235
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A E RN+RLGL +DGF P + + YSCWP+++ YNLPPD CM + L+ +IPGP
Sbjct: 236 AAEERNMRLGLSTDGFNPFNMKNTR-YSCWPVLLVNYNLPPDQCMKNENIMLSLLIPGPQ 294
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
P ID+YL+P I+DL LW G TYD K F ++A L WTI+ FP YG L+
Sbjct: 295 QPGNSIDVYLEPFIEDLNHLWKKGELTYDAFSKTTFTLKAMLFWTISDFPAYGNLAGCKV 354
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPN 299
G++ CP+C + TD+ L++ K F C R+ L H ++ NK A+ G+ E+
Sbjct: 355 KGKMGCPLCGKGTDSMWLKYSRKHV-FMCHRKGLAPTHSYK-NKKAWFDGKVEHGRKARI 412
Query: 300 LTSEQVWNKVKNKPKVQIVGGDASKPFGYGQKHN-------------------------- 333
LT +V + +KN + G+A K G +K N
Sbjct: 413 LTGREVSHNLKN---FKNDFGNA-KESGRKRKRNDCIESVTDSDNESSESEEDEEVEVDE 468
Query: 334 -----WTKRSIFWDLLYWKDNLLRHNLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKD 388
W KRSIF+ L YW++ +RHNLDVMH+E+N +I +T+++ GK+KD +RKD
Sbjct: 469 DELSRWKKRSIFFKLSYWENLPVRHNLDVMHVERNVGASIVSTLLHC-GKSKDGLNSRKD 527
Query: 389 IELYCRRKDLLLVEKSSGKFLK-PCANYTLPIEETKDVYRWVKELKMSD 436
++ RKD L + GK P A ++L E K + + E K D
Sbjct: 528 LQDLGVRKD--LHPNAQGKRTNLPAAPWSLSKSEKKTFCKHLFEFKGPD 574
>At4g07750 putative transposon protein
Length = 569
Score = 249 bits (635), Expect = 3e-66
Identities = 161/456 (35%), Positives = 233/456 (50%), Gaps = 53/456 (11%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+ +G ++I CV C LF N L C C R+K H K K VP+
Sbjct: 135 LKSFDMGYQKIHACVNDCCLFRKNY-----KKLDSCPKCNASRWKTNLHIGEIK-KGVPQ 188
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K L Y PIIPRL+ M+ S + + + WH N +++ +L HP D + W + K+P+F
Sbjct: 189 KVLRYFPIIPRLKGMFRSKEMDKDLRWHFSN---KSSDEKLCHPVDLVTWDQMNAKYPSF 245
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A E RN+R GL +DGF P + + YSCWP D+CM K + L +IPGP
Sbjct: 246 AGEERNLRHGLSTDGFNPFNMKNTR-YSCWP----------DLCMKKENIMLTLLIPGPQ 294
Query: 181 NPKVVIDIYLQPLIDDLKRLW-NGVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
P ID+YL+PLI+DL LW NG TYD K F ++A L+WTI+ FP YG L+
Sbjct: 295 QPGNNIDVYLEPLIEDLCHLWDNGELTYDAYSKSTFDLKAMLLWTISDFPAYGNLAGCKV 354
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPN 299
G++ CP+C ++TD+ L+ K + C + LP H FR K+ F G+ EN
Sbjct: 355 KGKMGCPLCGKNTDSMWLKFSRKHVYM-CHIKGLPPTHHFRMKKSWF-DGKAENGRKRRI 412
Query: 300 LTSEQVWNKVKN---------------KPKVQIVGG------------DASKPFGYGQKH 332
L+ ++++ +KN K V I D +
Sbjct: 413 LSGQEIYQNLKNFKNNFGTLKQSASKRKRTVNIEADSDSDDQSSESEEDEQDQVDEEELS 472
Query: 333 NWTKRSIFWDLLYWKDNLLRHNLDVMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELY 392
W K+SIF+ L YW++ L+RHNLDVMH+E+N +I +T+++ K+KD ARKD+E+
Sbjct: 473 RWKKKSIFFKLPYWQELLVRHNLDVMHVERNVAASIISTLLHC-WKSKDGLNARKDLEVL 531
Query: 393 CRRKDLLLVEKSSGKFLKPCANYTLPIEETKDVYRW 428
RKDL K +L P A ++L +E + RW
Sbjct: 532 GIRKDLHPKTKGKRTYL-PAALWSLS-KEIECTARW 565
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 239 bits (610), Expect = 3e-63
Identities = 168/518 (32%), Positives = 241/518 (46%), Gaps = 84/518 (16%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
+ G G I C C+L Y E+ ++D C C R+K KH+ K K +P
Sbjct: 127 LKMFGFGYDIIHACPNDCIL-YKKEYELRD----TCLGCSASRWKRDKHTGEEK-KGIPA 180
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
K L+Y I R +RM+ S + E + WH N + G +RHP D LAW + K P F
Sbjct: 181 KVLWYFSIKDRFKRMFRSKKMVEDLQWHFSN---ASVDGTMRHPIDSLAWAQVNDKWPQF 237
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A E RN+RLGL +D P + K YS WP+++ YN+ P MCM + L +I GP
Sbjct: 238 AAESRNLRLGLSTDKMNPFSIQNTK-YSTWPVLLVNYNMAPTMCMKAKNIMLTLLIHGPK 296
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
P ID+YL PLI+DLK L + G+ YD K+NF +RA L+W+IN +P T
Sbjct: 297 APSNNIDVYLAPLINDLKDLSSDGIQVYDSFFKENFTLRAMLLWSINDYP---------T 347
Query: 240 HGRLACPICMEDTDAFRLEHGGKTTWFGCTRRWLPDDHPFRRNKNAFMKGETENRGPPPN 299
G LA GC P RN
Sbjct: 348 LGTLA----------------------GCKDFKNDFGRPLERN----------------- 368
Query: 300 LTSEQVWNKVKNKPKVQIVGGDASKPFGYGQ---KHNWTKRSIFWDLLYWKDNLLRHNLD 356
NK K D + YG+ + W KRSI ++L YWKD +RHN+D
Sbjct: 369 -------NKRKRS--------DLCEDDEYGEDTHQWRWKKRSILFELPYWKDLPVRHNID 413
Query: 357 VMHIEKNFFDNIFNTVMNVKGKTKDNEKARKDIELYCRRKDLLLVEKSSGKFLKPCANYT 416
VMH+EKN D I +T+M K+KD ARK++E R +L + + +L A +
Sbjct: 414 VMHVEKNVSDVIVSTLMQ-SAKSKDGLNARKNLEDMGIRSNLHIQLRGKRTYLLHAA-FW 471
Query: 417 LPIEETKDVYRWVKELKMSDGYSSNLARCADVENGRMHGMKSHDTHIFMECLISTTFSAL 476
L +E K + + + DGY +N++ C V+ + G+KSHD H+ ++ L+ L
Sbjct: 472 LSKDEKKRFCKRLARFRGPDGYCANISNCVSVDPPVIGGLKSHDHHVLLQNLLPVALRGL 531
Query: 477 EPHVLNPLIEMSQ---YFKNSCSTTLREDDLIKMENEI 511
P P + +S+ YF C + + LI +E E+
Sbjct: 532 LPK--RPRVAISRLGHYFNRLCQRVIDPEKLISLEYEL 567
>At2g10630 En/Spm transposon hypothetical protein 1
Length = 515
Score = 231 bits (590), Expect = 6e-61
Identities = 115/240 (47%), Positives = 154/240 (63%), Gaps = 14/240 (5%)
Query: 1 MSKLGLGGKRIDCCVKGCMLFYDNEFGVKDGHLVECKFCQEPRYKGRKHSRSSKGKPVPR 60
M LGL ID C CMLF+ E K+ +C+FC R+K + R +K VP
Sbjct: 236 MRNLGLPYHTIDVCQNNCMLFWKEE--EKED---QCRFCGAKRWKPKDDRRRTK---VPY 287
Query: 61 KALFYLPIIPRLQRMYASMQTAEKMTWHSQNYERRNNSGELRHPSDGLAWKHFDQKHPNF 120
++YLPI RL+RMY S +TA M WH+ E ++ GE+ HPSD W++F HP F
Sbjct: 288 SCMWYLPIGDRLKRMYQSHKTAAAMRWHA---EHQSKEGEMNHPSDAAEWRYFQGLHPQF 344
Query: 121 ALEPRNVRLGLCSDGFTPHIQGSGKPYSCWPIIVTPYNLPPDMCMSKPYMFLAAVIPGPN 180
A +PRNV LGLC+DGF P G + +S WP+I+TPYNLPP MCM+ Y+FL + GPN
Sbjct: 345 AEDPRNVYLGLCTDGFNPF--GMSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPN 402
Query: 181 NPKVVIDIYLQPLIDDLKRLWN-GVHTYDISRKQNFVMRAALMWTINGFPTYGMLSEWGT 239
+P+ +D++LQPLI++LK LW+ GV YD+S QNF ++A L+WTI+ FP Y MLS W T
Sbjct: 403 HPRASLDVFLQPLIEELKELWSTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTT 462
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.139 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,731,106
Number of Sequences: 26719
Number of extensions: 659431
Number of successful extensions: 1512
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1324
Number of HSP's gapped (non-prelim): 57
length of query: 521
length of database: 11,318,596
effective HSP length: 104
effective length of query: 417
effective length of database: 8,539,820
effective search space: 3561104940
effective search space used: 3561104940
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC138465.9


