

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138465.7 + phase: 0 /pseudo
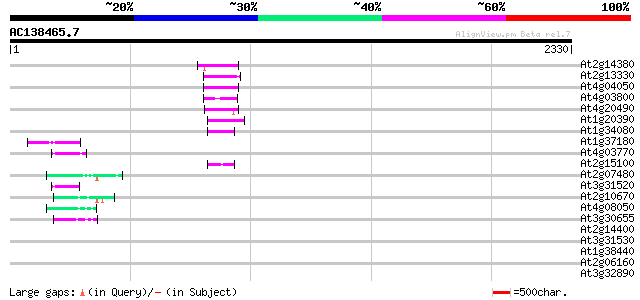
(2330 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g14380 putative retroelement pol polyprotein 76 2e-13
At2g13330 F14O4.9 73 2e-12
At4g04050 putative transposon protein 72 3e-12
At4g03800 71 9e-12
At4g20490 putative protein 69 2e-11
At1g20390 hypothetical protein 69 4e-11
At1g34080 putative protein 62 3e-09
At1g37180 hypothetical protein 52 3e-06
At4g03770 hypothetical protein 50 1e-05
At2g15100 putative retroelement pol polyprotein 50 1e-05
At2g07480 F9A16.15 48 8e-05
At3g31520 hypothetical protein 46 3e-04
At2g10670 pseudogene 46 3e-04
At4g08050 45 5e-04
At3g30655 hypothetical protein, 3' partial 45 7e-04
At2g14400 putative retroelement pol polyprotein 44 0.001
At3g31530 hypothetical protein 42 0.004
At1g38440 hypothetical protein 42 0.006
At2g06160 putative Athila retroelement ORF1 protein 41 0.007
At3g32890 Athila ORF 1, putative 41 0.009
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 76.3 bits (186), Expect = 2e-13
Identities = 52/195 (26%), Positives = 89/195 (44%), Gaps = 27/195 (13%)
Query: 780 EQAYVDHEVTVDRFGGIVGNITACNN---------------LW------------FSEDE 812
+Q + D V R I+G +TAC + LW F+ ++
Sbjct: 277 DQQHNDAPVPRRRVNMIMGGLTACRDSFRSIKEYIKSGAATLWSSPATKEMTPLTFTSED 336
Query: 813 MPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVK 872
+ HN L I ++ ++ +L+DTGSS+NV+ K L ++ + ++ S +
Sbjct: 337 LFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLT 396
Query: 873 AFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKL 932
FDG+ G I LP+ +G F V+D Y+ +LG PWIHD A+ S+ H+ +
Sbjct: 397 GFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCI 456
Query: 933 KFVKNGKLVTVHGEE 947
K + + T+ G +
Sbjct: 457 KIPTSIGIETIRGNQ 471
>At2g13330 F14O4.9
Length = 889
Score = 73.2 bits (178), Expect = 2e-12
Identities = 49/156 (31%), Positives = 78/156 (49%), Gaps = 6/156 (3%)
Query: 804 NNLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETP 863
+ + F E E+ + K H+ AL I ++ + LS ++VDTGSS++V+ + + ++
Sbjct: 45 DGITFWESEITDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSK 104
Query: 864 LRRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGA 923
L+ + F G +G I LP L F V+D A ++ +LGRPW+H A
Sbjct: 105 LQGRKTPLTGFAGDTTFSIGTIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKA 164
Query: 924 VTSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSC 959
V ST H+ +KF + V+G SQ SS C
Sbjct: 165 VPSTYHQCIKFPSYKGIAVVYG------SQRSSRKC 194
>At4g04050 putative transposon protein
Length = 681
Score = 72.4 bits (176), Expect = 3e-12
Identities = 43/145 (29%), Positives = 75/145 (51%)
Query: 804 NNLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETP 863
+ + F E E + K H+ AL I ++ + LS++++DTGSS++V+ ++ + ++
Sbjct: 519 HQITFWESETTDLNKPHDDALVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRMGHLDSE 578
Query: 864 LRRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGA 923
L+ + F G LG I LP L +F V++ A ++ +LGRPW+H A
Sbjct: 579 LQGRKTPLTGFAGDTTFSLGTIQLPTIARGVRRLTSFLVVNKKAPFNAILGRPWLHAMKA 638
Query: 924 VTSTLHKKLKFVKNGKLVTVHGEEA 948
V ST H+ +KF + + V A
Sbjct: 639 VPSTYHQCIKFPSDKGIAVVSAANA 663
>At4g03800
Length = 637
Score = 70.9 bits (172), Expect = 9e-12
Identities = 47/147 (31%), Positives = 75/147 (50%), Gaps = 20/147 (13%)
Query: 803 CNNLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVM---PKSTLDQLSY 859
C+++ F E+E + H+ AL I+++ + +S +LVDTGSS++++ P S L
Sbjct: 182 CSSVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLIFLGPPSPL----- 236
Query: 860 RETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIH 919
AF LG I LP+ G + ++ F V D A+Y+ +LG PWI
Sbjct: 237 ------------VAFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIF 284
Query: 920 DAGAVTSTLHKKLKFVKNGKLVTVHGE 946
AV ST H+ LKF + + T+ G+
Sbjct: 285 QMKAVPSTYHQWLKFPTSNGVETIWGD 311
>At4g20490 putative protein
Length = 853
Score = 69.3 bits (168), Expect = 2e-11
Identities = 44/155 (28%), Positives = 73/155 (46%), Gaps = 14/155 (9%)
Query: 808 FSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRS 867
F+ ++ + HN AL I + + + +L+DTGSS+N+M TL ++ E+ +
Sbjct: 465 FTSEDASGIQHPHNDALVIKLVMEDFDVERILIDTGSSVNLMFLKTLFKMGISESKIMPK 524
Query: 868 TFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 927
+ +DG K +GEI L + +G T F V+D Y+ +LG PWI+ A+ ST
Sbjct: 525 IRPMTGYDGEAKMSIGEIKLQVQVGGITQKTKFVVIDSEPIYNAILGSPWIYSMKAIPST 584
Query: 928 --------------LHKKLKFVKNGKLVTVHGEEA 948
L + + K G L+ + G A
Sbjct: 585 NLHTIRQPEGLTDMLRDREEAAKEGNLIAIKGRRA 619
>At1g20390 hypothetical protein
Length = 1791
Score = 68.6 bits (166), Expect = 4e-11
Identities = 43/164 (26%), Positives = 78/164 (47%), Gaps = 7/164 (4%)
Query: 820 HNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVKAFDGSRK 879
HN L + + ++ VL+DTGSS++++ K L ++ + ++ + + FDG
Sbjct: 567 HNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFV 626
Query: 880 NVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKFVKNGK 939
+G I LP+ +G + F V+ A Y+ +LG PWIH A+ ST H+ +KF +
Sbjct: 627 MTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPTHNG 686
Query: 940 LVTVHG-------EEAYLVSQLSSFSCIEAGSAEGTAFQGLTIE 976
+ T+ +Y S+L + ++ T G+ E
Sbjct: 687 IFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 730
Score = 39.7 bits (91), Expect = 0.021
Identities = 34/172 (19%), Positives = 70/172 (39%), Gaps = 4/172 (2%)
Query: 181 EFDRYNGLTCPQNHIIKY---VRKMGNYSDN-DSLMIHCFQDSLMEDAAEWYTSLSKDDV 236
+ + YNG P+ + + + + DN D+ F + L A W++ L + +
Sbjct: 173 KLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAHNWFSRLKPNSI 232
Query: 237 HTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEE 296
+F +L ++F HY + + L S+SQ ES R + ++ IT +
Sbjct: 233 DSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFKLVVTNITVPDEAAI 292
Query: 297 MTQTFLKTLKKDYVERMIIAAPNNFFEMVTMGTRLEEAVREGIIVFEKVESS 348
+ + + + + AP+ + + +R E E +I+ K S+
Sbjct: 293 VALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELEEEKLILARKHNST 344
>At1g34080 putative protein
Length = 811
Score = 62.4 bits (150), Expect = 3e-09
Identities = 36/112 (32%), Positives = 64/112 (57%)
Query: 823 ALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVKAFDGSRKNVL 882
AL IS++ + + +L+DTGSS++++ TL ++ + ++ + + +F L
Sbjct: 476 ALVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKDIKGAPSPLVSFTIETSMSL 535
Query: 883 GEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKF 934
G I LP+T +I F V D A+Y+ +LG PW+++ AV ST H+ +KF
Sbjct: 536 GTITLPVTAQGVVKMIEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQCVKF 587
>At1g37180 hypothetical protein
Length = 661
Score = 52.4 bits (124), Expect = 3e-06
Identities = 52/235 (22%), Positives = 96/235 (40%), Gaps = 22/235 (9%)
Query: 74 PSTAAGTSGTANN-------GAVPGTNTISINTTLPQTTAT---VTEPLVHAISQGVNIN 123
PS GT+ T N G +N T +P A +++P + +
Sbjct: 205 PSVEVGTTSTQNPTTSSSPAGFAAVSNETIARTLVPLDPAIRLPISDPRQEPSAVNSELQ 264
Query: 124 AHHGSIPVIKTMEERMEELAKELRREIKANRGNGDSVKTHDLCLVPKVDVPKKFKIPEFD 183
+ I + + A E+ + I+A R + + L ++ + FK+P
Sbjct: 265 SLQDQIWAMNAKVHQATTSAPEVEKVIEATRRTPFTPRISKL----RIREFRDFKLPV-- 318
Query: 184 RYNGLTCPQNHIIKY---VRKMG-NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTF 239
YNG P H+ + VR++ + D+ + F ++L A W+T L + + F
Sbjct: 319 -YNGKGDPNEHLTSFQVIVRRVPLEPYEEDAGLCKLFSENLSGPALTWFTQLEEGSIDNF 377
Query: 240 DELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDE 294
+L+ AF YG+ + L +LSQ D+ R Y +++ +I P+L +
Sbjct: 378 KQLSTAFIKQYGYFIKSDITEAHLWNLSQSADDPLRTYIIVFKEIMVQI-PSLSD 431
>At4g03770 hypothetical protein
Length = 464
Score = 50.4 bits (119), Expect = 1e-05
Identities = 42/148 (28%), Positives = 63/148 (42%), Gaps = 12/148 (8%)
Query: 174 PKKFKIPEFDRYNGLTCPQNH----IIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYT 229
P K +I F NG + P+ H II R + D+ + H F + L A W+T
Sbjct: 231 PGKLRIEYF---NGSSDPKGHLKLFIISVARAKFRPEERDAGLCHLFVEHLKGPALNWFT 287
Query: 230 SLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARIT 289
L + V +F EL+ F Y + L SLSQ+ +E R++ +R A++
Sbjct: 288 RLKGNSVDSFQELSTLFLKQYSVLIDPSTSDADLWSLSQQPNEPLRDFLAKFRSTLAKVE 347
Query: 290 PALDEEEMTQTFLKTLKKDYVERMIIAA 317
D L TLKK R ++A
Sbjct: 348 GIND-----LAALSTLKKALCLRARLSA 370
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 50.4 bits (119), Expect = 1e-05
Identities = 35/116 (30%), Positives = 56/116 (48%), Gaps = 6/116 (5%)
Query: 823 ALHISVNW-KSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRR---STFLVKAFDGSR 878
ALH + NW ++ L S+ N PK+ + + T LR+ T + S
Sbjct: 292 ALHRATNWINAEEERAFLAKKFSASNAAPKAPQPATTKKPTELRKPAAGTCTTMLCETSM 351
Query: 879 KNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKF 934
LG + LP+T ++ F V D A+Y+ +LG PW+++ V ST H+ +KF
Sbjct: 352 S--LGTVVLPVTAQGVVKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKF 405
>At2g07480 F9A16.15
Length = 1012
Score = 47.8 bits (112), Expect = 8e-05
Identities = 87/383 (22%), Positives = 141/383 (36%), Gaps = 85/383 (22%)
Query: 151 KANRGNGDSVKTHD--LCLVPKVDVPKKFKIP-------EFDRYNGLTC--PQNHIIKYV 199
++N G GD+ + H +VP FKI + ++++GL P +H+ +
Sbjct: 6 QSNIGAGDAPRNHHQRAGIVPPPIQNNNFKIKSDLISMIQGNKFHGLPMEDPLDHLDNFD 65
Query: 200 RKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTR 255
R N +S + F SL + A W +L + T+D+ AF + + N+R
Sbjct: 66 RLCSLTKINGVSEESFKLRLFPFSLGDKAHLWEKTLPVKSIDTWDDCKKAFLAKFFSNSR 125
Query: 256 LKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEEMTQTFLKTLKKDYVERMII 315
R + +QK ESF E + ++G + P + + L+ L K RM++
Sbjct: 126 KARLRSEISGFNQKNSESFSEAWERFKGYTTQ-CPHHESLPPQYSILRCLPK---IRMLL 181
Query: 316 AAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNASKRYGNDH-------------HKK 362
+N G L + V EG +E VE+ + + Y D+ HKK
Sbjct: 182 DTASN-------GNILNKDVAEG---WELVENLAQSHRNYNEDYDRTNRGSSDSEDKHKK 231
Query: 363 K-----------------------ETEVGMVSAGAGQSMATVAPINAA--------QLPP 391
+ E E+ + G ++ V+ + P
Sbjct: 232 EIKTLNDRIDKLVLAQQRNVYYITEEELTQLQDGENLTIEEVSYLQNQGGYNKGFNNYKP 291
Query: 392 SYPYAPYSQHPFFPPFYHQYPLPSGQPQ-----VPVNAVVQQMQQ-QPPVQQQQHQQARP 445
+P Y + P YP P QP VP N Q Q PP QQ QQ
Sbjct: 292 PHPNLSYRSNNVANPQDQVYP-PQNQPTQAKPFVPYNQGYNQKQNFGPPGFTQQPQQ--- 347
Query: 446 TFPPIPMLYAELLPTLLLRGHCT 468
P + LP L++GH +
Sbjct: 348 --PSAQDSEMKTLPQQLVQGHAS 368
>At3g31520 hypothetical protein
Length = 528
Score = 45.8 bits (107), Expect = 3e-04
Identities = 33/119 (27%), Positives = 53/119 (43%), Gaps = 7/119 (5%)
Query: 174 PKKFKIPEFDRYNGLTCPQNH----IIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYT 229
P K +I F NG + P+ H II R + D+ + H F + L A W++
Sbjct: 239 PGKLRIEYF---NGSSDPKRHLKSFIISVARTKFRPEERDAGLCHLFVEHLKGPALCWFS 295
Query: 230 SLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARI 288
L + + +F EL+ F Y L + L SLSQ+ +E R++ R A++
Sbjct: 296 RLEGNSMDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLRDFLAKLRSTLAKV 354
>At2g10670 pseudogene
Length = 929
Score = 45.8 bits (107), Expect = 3e-04
Identities = 70/312 (22%), Positives = 117/312 (37%), Gaps = 72/312 (23%)
Query: 183 DRYNGLTC--PQNHIIKYVRKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDV 236
++++GL P +H+ + R G N D + F SL + A W +L K+ +
Sbjct: 84 NKFHGLPMEDPLDHLDELERLCGLTKINGVSEDGFKLRLFPFSLGDKAHLWEKTLPKNSI 143
Query: 237 HTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEE 296
T+D+ AF + + N+R R + + K++ESF E + ++G T
Sbjct: 144 TTWDDCKMAFLAKFFSNSRTARLRNEISGFTLKQNESFCEAWERFKGYQ---TKCPHHGF 200
Query: 297 MTQTFLKTLKKDYVE--RMIIAAPNNFFEMVTMGTRLEEAVREGIIVFEKV-ESSVNASK 353
+ L TL + + RM++ +N G L++ + EG + E + +S N ++
Sbjct: 201 SQASLLNTLYRGVLPKIRMLLDTASN-------GNFLKKDIEEGWELVENLAQSGGNYNE 253
Query: 354 RYGND----------HHKK----------------------KETEVGMVSAGAGQSMATV 381
YG HH++ E E V G A +
Sbjct: 254 DYGRSIYTSSNTDDKHHREMKALNDKLDKIIQMQQKHVHFISEDEPFQVQEGENDQCAEI 313
Query: 382 A---------PINAAQLPPSYPYAPYSQHPFFPP------FYHQYPLPSG------QPQV 420
P AA P+ P+ PY+Q F P Y Q P G Q
Sbjct: 314 RYVHNQGPSLPSAAAAAEPTKPFVPYNQSLGFVPKQQFQGGYQQQQPPPGFTPHQQQAHA 373
Query: 421 PVNAVVQQMQQQ 432
P N+ + + QQ
Sbjct: 374 PQNSDIMTVLQQ 385
Score = 34.3 bits (77), Expect = 0.88
Identities = 33/115 (28%), Positives = 53/115 (45%), Gaps = 13/115 (11%)
Query: 836 SNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVKAFDGSRKNVLGEI-DLPMTIGHE 894
+N L D G+S+++MP S + +L + T ++ D + K G + D+P+ I
Sbjct: 589 NNCLCDLGASVSLMPLSVVKKLGFVHYKPCDLTLILA--DRTSKTPFGLLEDVPVMINGV 646
Query: 895 TFLITFQV--MDINASYSCLLGRPWIHDAGAVTSTLHKKLKFVKNGKLVTVHGEE 947
F V MD + +LGRP++ GAV VK GK+ GE+
Sbjct: 647 EVPTDFVVLEMDGESKDPLILGRPFLASVGAVID--------VKQGKINLNLGED 693
>At4g08050
Length = 1428
Score = 45.1 bits (105), Expect = 5e-04
Identities = 55/223 (24%), Positives = 90/223 (39%), Gaps = 28/223 (12%)
Query: 152 ANRGNGDSVKTHDLC--LVPKVDVPKKFKIP-------EFDRYNGLTC--PQNHIIKYVR 200
AN G GD+ + H + P F+I + ++++GL P +H+ ++ R
Sbjct: 12 ANIGAGDAPRDHRQRKGIAPPAIQNNNFEIKSGLISMIQGNKFHGLPMEDPLDHLDEFDR 71
Query: 201 KMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRL 256
N D + F SL + A W +LS D + T+D+ AF S + N R
Sbjct: 72 LCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLSHDSITTWDDYKKAFLSKFFSNART 131
Query: 257 KPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEEMTQ-TFLKTLKKDYVERMII 315
R + SQK ESF E + ++G T T+ + L TL + + R+
Sbjct: 132 ARLRNEIYGFSQKTGESFCEAWERFKG----YTNQCPHHSFTKASLLSTLYRGVLPRI-- 185
Query: 316 AAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNASKRYGND 358
+ + G + V EG +E VE+ + Y D
Sbjct: 186 ---RMLLDTASNGNFQNKDVEEG---WELVENLAQSDGNYNED 222
Score = 39.3 bits (90), Expect = 0.027
Identities = 32/102 (31%), Positives = 49/102 (47%), Gaps = 9/102 (8%)
Query: 836 SNVLVDTGSSLNVMPKS---TLDQLSYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIG 892
S+ L D G+S+N+MP S L+ + Y+ L L+ A SRK+ DLP+ I
Sbjct: 589 SDCLCDLGASVNLMPLSMARRLEFIQYKPCDLT----LILADRSSRKHFGMLKDLPVMIN 644
Query: 893 HETFLITFQVMDINASYS--CLLGRPWIHDAGAVTSTLHKKL 932
F V+D+ + +LGRP++ GAV K+
Sbjct: 645 GVEVPTDFVVLDMEVEHKDPLILGRPFLASVGAVIDVKEGKI 686
>At3g30655 hypothetical protein, 3' partial
Length = 660
Score = 44.7 bits (104), Expect = 7e-04
Identities = 43/189 (22%), Positives = 80/189 (41%), Gaps = 17/189 (8%)
Query: 183 DRYNGLTC--PQNHIIKYVRKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDV 236
++++GL P +H+ ++ R G N D + F SL + A W +L ++ +
Sbjct: 52 NKFHGLPMEDPLDHLDEFERLYGLTKINGVSEDGFKLRLFPFSLGDKAHLWEKTLPQNSI 111
Query: 237 HTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEE 296
+D+ AF + + N+R R + +QK++ESF E W T
Sbjct: 112 TAWDDCKKAFLAKFFSNSRTARLRNEISGFTQKQNESFGE---AWERFKGYQTKCPHHGF 168
Query: 297 MTQTFLKTLKKDYVERMIIAAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNASKRYG 356
+ L TL + + ++ I + + G L++ V EG +E VE+ + Y
Sbjct: 169 KQASLLSTLYRGVLPKIRI-----LLDTASNGNFLKKDVEEG---WELVENFAQSDGNYN 220
Query: 357 NDHHKKKET 365
D+ + T
Sbjct: 221 EDYDRSIRT 229
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 43.5 bits (101), Expect = 0.001
Identities = 26/99 (26%), Positives = 44/99 (44%), Gaps = 4/99 (4%)
Query: 183 DRYNGLTCPQNHIIKYVRKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHT 238
+ YNGL P+ ++ ++ G N +D D F ++L A W+T L +
Sbjct: 163 ESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSENLCGQALMWFTQLEPGSISN 222
Query: 239 FDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREY 277
F+EL+ F Y + L +LSQ +E+ R +
Sbjct: 223 FNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAF 261
Score = 37.4 bits (85), Expect = 0.10
Identities = 24/88 (27%), Positives = 47/88 (53%), Gaps = 4/88 (4%)
Query: 805 NLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPL 864
++ F E E + H+ AL ++++ + +S +L+DTGSS++++ STL+++ +
Sbjct: 408 SILFDEKETQHLERSHDDALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISRADV 467
Query: 865 RRSTFLVKAFDGSRKNVLGEIDLPMTIG 892
R V+ K V E+D + IG
Sbjct: 468 NRRKLGVE----RAKAVNDEVDKLLKIG 491
>At3g31530 hypothetical protein
Length = 831
Score = 42.0 bits (97), Expect = 0.004
Identities = 20/53 (37%), Positives = 29/53 (53%)
Query: 882 LGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKF 934
LG I LP+ T ++ F + D Y+ ++G PW++ AV ST H LKF
Sbjct: 3 LGTIKLPVRAKGVTKIVDFSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKF 55
>At1g38440 hypothetical protein
Length = 263
Score = 41.6 bits (96), Expect = 0.006
Identities = 32/105 (30%), Positives = 46/105 (43%), Gaps = 12/105 (11%)
Query: 179 IPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHT 238
+ EFDR LT K+ S+ D + F SL + A W +L D + T
Sbjct: 19 LDEFDRLYNLT-----------KINGVSE-DGFKLRLFPFSLGDKAHIWEKNLPHDSITT 66
Query: 239 FDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRG 283
+D+ AF S + N R R + SQK ESF E + ++G
Sbjct: 67 WDDCKKAFLSKFFSNARTARLRNEISGFSQKTSESFCEAWERFKG 111
>At2g06160 putative Athila retroelement ORF1 protein
Length = 750
Score = 41.2 bits (95), Expect = 0.007
Identities = 53/245 (21%), Positives = 98/245 (39%), Gaps = 29/245 (11%)
Query: 121 NINAHHGSIPVIKTMEERMEELAKELRREIKANRGNGDSVKTHDLCLVPKVDVPKKFKIP 180
N N HG V+ ++ E+ L ++ N K H L + +D +
Sbjct: 22 NHNQRHGI--VLPLVQNNNFEIKSGLIAMVQGN-------KFHGLPMEDSLD-----HLD 67
Query: 181 EFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFD 240
EF+R LT K+ S+ D F SL + W +L ++ + T+D
Sbjct: 68 EFERLCDLT-----------KINGVSE-DGFKFRLFPFSLGDKTHLWEKTLPQNSITTWD 115
Query: 241 ELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARIT-PALDEEEMTQ 299
+ AF + + N+R R + +QK++ESF E + ++G + P + +
Sbjct: 116 DCKKAFFAKFFSNSRTARLRNEISGFTQKQNESFCEAWERFKGYQTKCPHPGFKQASLLS 175
Query: 300 TFLKTLKKDYVERMIIAAPNNFFEM-VTMGTRLEEAVREGIIVF-EKVESSVNASKRYGN 357
T + + + A+ NF V G L E + + + E + S++ S +
Sbjct: 176 TLYRGVLPKLRMLLDTASNGNFLNKDVEKGWELVENLAQSDGNYNENYDKSISTSSESDD 235
Query: 358 DHHKK 362
HH++
Sbjct: 236 KHHRE 240
>At3g32890 Athila ORF 1, putative
Length = 755
Score = 40.8 bits (94), Expect = 0.009
Identities = 32/105 (30%), Positives = 46/105 (43%), Gaps = 12/105 (11%)
Query: 179 IPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHT 238
+ EFDR LT K+ S+ D + F SL + A W +L D + T
Sbjct: 19 LDEFDRLYNLT-----------KINGVSE-DGFKLRLFPFSLGDKAHIWEKNLPHDSITT 66
Query: 239 FDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRG 283
+D+ AF S + N R R + SQK ESF E + ++G
Sbjct: 67 WDDCKKAFLSKFFSNARTARLRNEISGFSQKTRESFCEAWERFKG 111
Score = 34.3 bits (77), Expect = 0.88
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Query: 836 SNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVKAFDGSRKNVLGEI-DLPMTIGHE 894
S+ L D G+S+++MP S +L + + T ++ D S + G + DLP+ I
Sbjct: 541 SDCLCDLGASVSLMPLSVARRLEFIQYKPCDLTLILA--DRSFRKPFGMLKDLPVMINGV 598
Query: 895 TFLITFQVMDINASYS--CLLGRPWIHDAGAVTSTLHKKL 932
F V+D+ + +LGRP + GAV K+
Sbjct: 599 EVPTDFVVLDMEVEHKDPLILGRPLLASVGAVIDVREGKI 638
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.338 0.146 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,598,106
Number of Sequences: 26719
Number of extensions: 1959494
Number of successful extensions: 8226
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 7908
Number of HSP's gapped (non-prelim): 298
length of query: 2330
length of database: 11,318,596
effective HSP length: 115
effective length of query: 2215
effective length of database: 8,245,911
effective search space: 18264692865
effective search space used: 18264692865
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC138465.7


