

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.6 + phase: 0
(250 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
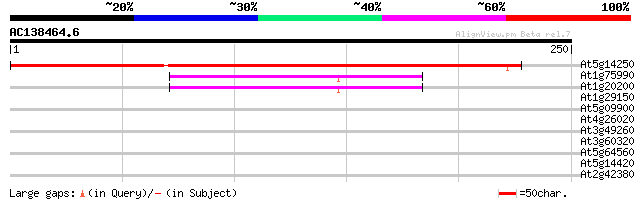
Score E
Sequences producing significant alignments: (bits) Value
At5g14250 unknown protein 292 1e-79
At1g75990 proteasome regulatory subunit S3 like protein 47 1e-05
At1g20200 proteasome regulatory subunit S3 like protein 43 2e-04
At1g29150 19S proteosome subunit 9 like protein 30 0.93
At5g09900 26S proteasome p55 protein-like 30 1.6
At4g26020 hypothetical protein 28 3.5
At3g49260 SF16 -like protein 28 3.5
At3g60320 bZIP like protein 28 6.0
At5g64560 unknown protein 27 7.9
At5g14420 unknown protein 27 7.9
At2g42380 bZip transcription factor AtbZip34 27 7.9
>At5g14250 unknown protein
Length = 429
Score = 292 bits (748), Expect = 1e-79
Identities = 156/230 (67%), Positives = 178/230 (76%), Gaps = 3/230 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MICIG KRFQKAL+LL+NVVTAPM +NAI +EAYKKYILVSLI +GQF+ +LPK AS
Sbjct: 189 MICIGLKRFQKALELLYNVVTAPMHQVNAIALEAYKKYILVSLIHNGQFTNTLPKCASTA 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQR+ K + P IEL YN+GK+ E+EA V A FE D NLGL KQ VSS+YKRNI
Sbjct: 249 AQRSFKNYT-GPYIELGNCYNDGKIGELEALVVARNAEFEEDKNLGLVKQAVSSLYKRNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
RLTQ YLTLSL+DIAN V L +AKEAEMHVLQMIQDG+I+A INQKDGMVRFLEDPEQY
Sbjct: 308 LRLTQKYLTLSLQDIANMVQLGNAKEAEMHVLQMIQDGQIHALINQKDGMVRFLEDPEQY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKV--SVQSFSFG 228
K+ EMIE +DS IQR + LSK L A DE +SCD LYL KV Q + FG
Sbjct: 368 KSSEMIEIMDSVIQRTIGLSKNLLAMDESLSCDPLYLGKVGRERQRYDFG 417
>At1g75990 proteasome regulatory subunit S3 like protein
Length = 487
Score = 46.6 bits (109), Expect = 1e-05
Identities = 33/116 (28%), Positives = 55/116 (46%), Gaps = 3/116 (2%)
Query: 72 PCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNIQRLTQTYLTLS 131
P EL G + A F D L ++ ++ + ++ ++ +Y +S
Sbjct: 311 PYFELTNAVRIGDLELFGKIQEKFAKTFAEDRTHNLIVRLRHNVIRTGLRNISISYSRIS 370
Query: 132 LEDIANTVHLNSAK---EAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQYKTCE 184
L+D+A + LNSA +AE V + I+DG I ATI+ K+G + E + Y T E
Sbjct: 371 LQDVAQKLRLNSANPVADAESIVAKAIRDGAIDATIDHKNGCMVSKETGDIYSTNE 426
>At1g20200 proteasome regulatory subunit S3 like protein
Length = 488
Score = 42.7 bits (99), Expect = 2e-04
Identities = 31/116 (26%), Positives = 52/116 (44%), Gaps = 3/116 (2%)
Query: 72 PCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNIQRLTQTYLTLS 131
P EL G + F D L ++ ++ + ++ ++ +Y +S
Sbjct: 312 PYFELTNAVRIGDLELFRTVQEKFLDTFAQDRTHNLIVRLRHNVIRTGLRNISISYSRIS 371
Query: 132 LEDIANTVHLNSAK---EAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQYKTCE 184
L D+A + LNS +AE V + I+DG I ATI+ K+G + E + Y T E
Sbjct: 372 LPDVAKKLRLNSENPVADAESIVAKAIRDGAIDATIDHKNGCMVSKETGDIYSTNE 427
>At1g29150 19S proteosome subunit 9 like protein
Length = 419
Score = 30.4 bits (67), Expect = 0.93
Identities = 25/100 (25%), Positives = 46/100 (46%), Gaps = 9/100 (9%)
Query: 112 VSSMY----KRNIQRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQK 167
+SS+Y ++N+ RL + + + + IA + L E + QMI D + T++Q
Sbjct: 319 LSSLYDTLLEQNLCRLIEPFSRVEIAHIAELIGL-PLDHVEKKLSQMILDKKFAGTLDQG 377
Query: 168 DGMVRFLEDPE----QYKTCEMIEHIDSSIQRIMALSKKL 203
G + EDP+ T E I ++ + + S K+
Sbjct: 378 AGCLIIFEDPKADAIYSATLETIANMGKVVDSLYVRSAKI 417
>At5g09900 26S proteasome p55 protein-like
Length = 442
Score = 29.6 bits (65), Expect = 1.6
Identities = 17/57 (29%), Positives = 34/57 (58%), Gaps = 1/57 (1%)
Query: 117 KRNIQRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRF 173
+ NI +++ Y ++L+ +A + L S +EAE H+ +M+ + A I++ G+V F
Sbjct: 343 EHNILVVSKYYARITLKRLAELLCL-SMEEAEKHLSEMVVSKALIAKIDRPSGIVCF 398
>At4g26020 hypothetical protein
Length = 273
Score = 28.5 bits (62), Expect = 3.5
Identities = 31/164 (18%), Positives = 69/164 (41%), Gaps = 16/164 (9%)
Query: 54 PKYASPLAQRNLKLFCQNPCIELAQTYNNG---KVAEVEAFVNANAGRFEADNNLGLAKQ 110
P P RNL++ + CI L ++ + K+ E +N + +++ +
Sbjct: 43 PSLIEPNELRNLEILISSCCISLTGSFRDDLDFKIGLNE--INVYRSKSNVESSRREVLE 100
Query: 111 VVSSMYKRNIQRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGE--IYATINQKD 168
+ + K +RL + Y T SL + A+ H N+ + + +++ E I + + +K
Sbjct: 101 ISNKNLKEENERLKKLY-TESLNNFADQEHRNALESLRQKHVTKVEELEYKIRSLLVEKA 159
Query: 169 GMVRFLEDPEQYKTCEMIEHIDSSIQRIMALSKKLTATDEQISC 212
++ + + + ++ I A+SKKL ++ C
Sbjct: 160 TNDMVID--------RLRQDLTANKSHIQAMSKKLDRVVTEVEC 195
>At3g49260 SF16 -like protein
Length = 471
Score = 28.5 bits (62), Expect = 3.5
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 4/64 (6%)
Query: 20 VTAPMTMMNAIVIEAYKKYILVSLIRHG----QFSTSLPKYASPLAQRNLKLFCQNPCIE 75
VT P ++ I+ ++YI + R G + T +P Y +P A K+ Q P ++
Sbjct: 340 VTTPTSLKGNIIGLIDREYIDLGSYRQGHKQRKSPTHIPSYMAPTASAKAKVRDQGPTVK 399
Query: 76 LAQT 79
L T
Sbjct: 400 LQGT 403
>At3g60320 bZIP like protein
Length = 796
Score = 27.7 bits (60), Expect = 6.0
Identities = 18/71 (25%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Query: 116 YKRNIQRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLE 175
+K + R+ T + +++ N VH+ SAK+A+ H ++ + + +K VR LE
Sbjct: 638 WKLALDRIPDTVASEAIKSFINVVHVISAKQADEHKIKK-RTESASKELEKKASSVRNLE 696
Query: 176 DPEQYKTCEMI 186
+ Y++ M+
Sbjct: 697 -RKYYQSYSMV 706
>At5g64560 unknown protein
Length = 394
Score = 27.3 bits (59), Expect = 7.9
Identities = 16/63 (25%), Positives = 31/63 (48%)
Query: 138 TVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQYKTCEMIEHIDSSIQRIM 197
TVH + E+ +L +I +T+N+ + +++D E Y ++ H + IQ +
Sbjct: 270 TVHGDENDVEELEMLLEAYFMQIDSTLNRLTTLREYIDDTEDYINIQLDNHRNQLIQLEL 329
Query: 198 ALS 200
LS
Sbjct: 330 VLS 332
>At5g14420 unknown protein
Length = 468
Score = 27.3 bits (59), Expect = 7.9
Identities = 19/88 (21%), Positives = 40/88 (44%), Gaps = 4/88 (4%)
Query: 99 FEADNNLGLAKQVVSSMYKRNIQRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDG 158
F +++ + V S YK + +L T I + + + HVL +I DG
Sbjct: 194 FNSEDRFCNGFEEVLSRYKEIVPQLKLAGPTSFAPIIDMAMTIVEQSGGQYHVLVIIADG 253
Query: 159 EIYATINQKDGMVRFLEDPEQYKTCEMI 186
++ +++ ++G + P++ KT + I
Sbjct: 254 QVTRSVDTENGQL----SPQEQKTVDAI 277
>At2g42380 bZip transcription factor AtbZip34
Length = 300
Score = 27.3 bits (59), Expect = 7.9
Identities = 14/37 (37%), Positives = 19/37 (50%)
Query: 107 LAKQVVSSMYKRNIQRLTQTYLTLSLEDIANTVHLNS 143
L K KR I+RL Q Y SL ++ N HL++
Sbjct: 240 LFKDAHQEALKREIERLRQVYNQQSLTNVENANHLSA 276
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,756,929
Number of Sequences: 26719
Number of extensions: 172776
Number of successful extensions: 523
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 514
Number of HSP's gapped (non-prelim): 11
length of query: 250
length of database: 11,318,596
effective HSP length: 97
effective length of query: 153
effective length of database: 8,726,853
effective search space: 1335208509
effective search space used: 1335208509
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC138464.6


