

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.5 + phase: 0
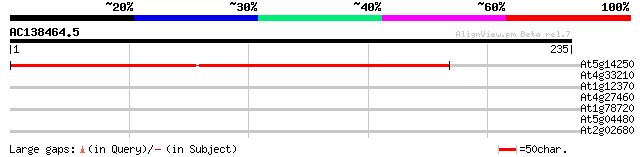
(235 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g14250 unknown protein 182 1e-46
At4g33210 unknown protein 31 0.50
At1g12370 putative type II CPD photolyase 31 0.50
At4g27460 unknown protein 28 5.5
At1g78720 hypothetical protein 28 5.5
At5g04480 unknown protein 27 9.4
At2g02680 hypothetical protein 27 9.4
>At5g14250 unknown protein
Length = 429
Score = 182 bits (463), Expect = 1e-46
Identities = 92/184 (50%), Positives = 126/184 (68%), Gaps = 1/184 (0%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY 60
++ +E +I IQGLS S D++ LH +L+ + ++LR++ + L +D S HSLG+LY
Sbjct: 5 VNSVEAVITSIQGLSGSPEDLSALHDLLRGAQDSLRAEPGVNFSTLDQLDASKHSLGYLY 64
Query: 61 LLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPI 120
L+ + VS ++ A E +PIIARFIN+C QIRLA KFVS+CK LKD V L P+
Sbjct: 65 FLEVLTCGPVSKEKAAYE-IPIIARFINSCDAGQIRLASYKFVSLCKILKDHVIALGDPL 123
Query: 121 RGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLY 180
RGV P+ A++KLQVS++ LT LH + L LCL AK YK+G SIL DD+ E+D PRD +LY
Sbjct: 124 RGVGPLLNAVQKLQVSSKRLTALHPDVLQLCLQAKSYKSGFSILSDDIVEIDQPRDFFLY 183
Query: 181 CYYG 184
YYG
Sbjct: 184 SYYG 187
>At4g33210 unknown protein
Length = 990
Score = 31.2 bits (69), Expect = 0.50
Identities = 38/153 (24%), Positives = 63/153 (40%), Gaps = 11/153 (7%)
Query: 32 DETLRSDSSRLYPILQLIDPSIHSL-GFLYLLDAFSAASVSNQQQAEEAVPIIARFINAC 90
D+ SD+ + L D +H + FL +D +A V Q + A R +N
Sbjct: 180 DDNGSSDTEDFEVHIDLTDDLLHMVFSFLNHVDLCRSAMVCRQWRVASAHEDFWRVLN-- 237
Query: 91 SVEQIRLAPEKFVSVCKRLKDL--VTLLEAPIRGVAPM--FTALRKLQV---SAEHLTPL 143
E IR++ E+F ++C R + V + AP M T LR L+V H++
Sbjct: 238 -FENIRISMEQFENMCSRYPNATEVNVYGAPAVNALAMKAATTLRNLEVLTIGKGHISES 296
Query: 144 HAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRD 176
+ L C + + +IL + E+ D
Sbjct: 297 FFQALGECNMLRSVTVSDAILGNGAQEIHLSHD 329
>At1g12370 putative type II CPD photolyase
Length = 490
Score = 31.2 bits (69), Expect = 0.50
Identities = 26/106 (24%), Positives = 42/106 (39%), Gaps = 18/106 (16%)
Query: 47 QLIDPSIHSLGFL--------YLLDAFSAASVSNQQQAEEAVPIIARFINACSVEQI--R 96
Q +D LGF+ + +D+ Q A+E +P F+ C +
Sbjct: 69 QFLDAKARQLGFMLKGLRQLHHQIDSLQIPFFLLQGDAKETIP---NFLTECGASHLVTD 125
Query: 97 LAPEKFVSVCK-----RLKDLVTLLEAPIRGVAPMFTALRKLQVSA 137
+P + + CK R D + + E V PM+ A KL+ SA
Sbjct: 126 FSPLREIRRCKDEVVKRTSDSLAIHEVDAHNVVPMWAASSKLEYSA 171
>At4g27460 unknown protein
Length = 391
Score = 27.7 bits (60), Expect = 5.5
Identities = 24/121 (19%), Positives = 47/121 (38%), Gaps = 6/121 (4%)
Query: 2 DPLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYP---ILQLIDPSIHS--- 55
DP LI +I ++ + D + S L + P ++Q++ +
Sbjct: 230 DPFTSLIGEISPMTLTCCDETAAAAVATLSAGDLMAYIDGANPPESLVQIVRNRLEDKGL 289
Query: 56 LGFLYLLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTL 115
+G + L D+ S+ S S+ +EE P+ R+A + VC L+ +
Sbjct: 290 IGLMSLFDSLSSYSTSSGYSSEEEAPVRTTSYGRSMSSSARMARKSEAIVCNPKSSLMAV 349
Query: 116 L 116
+
Sbjct: 350 M 350
>At1g78720 hypothetical protein
Length = 475
Score = 27.7 bits (60), Expect = 5.5
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Query: 119 PIRGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKC 156
P+ G+A TA L A H P HA F ++ +LA C
Sbjct: 338 PVGGIAYYITAPSSLAEMATH--PFHALFYLVFMLAAC 373
>At5g04480 unknown protein
Length = 1050
Score = 26.9 bits (58), Expect = 9.4
Identities = 16/48 (33%), Positives = 27/48 (55%), Gaps = 4/48 (8%)
Query: 159 TGLSILDDDVFEV----DHPRDLYLYCYYGCVAFFFSFELEKRDRCRI 202
T L +D+D+ E DHPR+ +L+ G V + +E E+ +R R+
Sbjct: 976 TLLKSMDEDLAEAADDKDHPRERWLWPLTGEVHWKGVYEREREERYRL 1023
>At2g02680 hypothetical protein
Length = 649
Score = 26.9 bits (58), Expect = 9.4
Identities = 17/72 (23%), Positives = 29/72 (39%), Gaps = 14/72 (19%)
Query: 176 DLYLY--CYYGCVAFFFS-----------FELEKRDRCRICSYISVLHFSYI-TYCNFFN 221
D+Y++ C + FF F + C IC + LHF+Y+ C+F
Sbjct: 211 DMYMHTLCAQAVIPFFIDQPKRHAHILTLFPRQASLTCNICGVLDELHFTYVCPICDFVT 270
Query: 222 ILDLFFTSFSIK 233
D + +I+
Sbjct: 271 HADCIYIPQTIR 282
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.329 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,995,586
Number of Sequences: 26719
Number of extensions: 198394
Number of successful extensions: 640
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 638
Number of HSP's gapped (non-prelim): 7
length of query: 235
length of database: 11,318,596
effective HSP length: 96
effective length of query: 139
effective length of database: 8,753,572
effective search space: 1216746508
effective search space used: 1216746508
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC138464.5


