

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138452.7 + phase: 0 /pseudo
(677 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
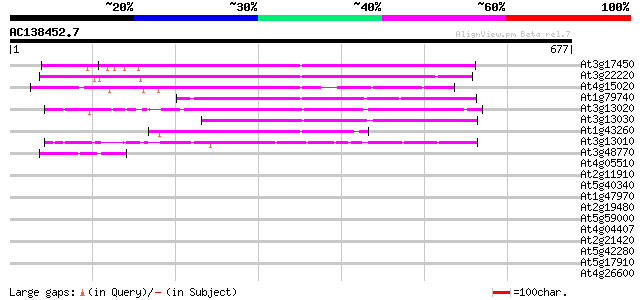
Score E
Sequences producing significant alignments: (bits) Value
At3g17450 unknown protein 311 1e-84
At3g22220 hypothetical protein 254 1e-67
At4g15020 putative protein 202 6e-52
At1g79740 hypothetical protein 191 1e-48
At3g13020 hypothetical protein 190 3e-48
At3g13030 unknown protein (At3g13030) 184 1e-46
At1g43260 hypothetical protein 153 3e-37
At3g13010 hypothetical protein 150 2e-36
At3g48770 hypothetical protein 44 3e-04
At4g05510 39 0.007
At2g11910 unknown protein 35 0.11
At5g40340 unknown protein 33 0.40
At1g47970 Unknown protein (T2J15.12) 33 0.52
At2g19480 putative nucleosome assembly protein 33 0.68
At5g59000 unknown protein 32 0.89
At4g04407 putative protein 32 0.89
At2g21420 Mutator-like transposase 32 0.89
At5g42280 putative protein 32 1.2
At5g17910 putative protein 32 1.2
At4g26600 unknown protein 32 1.2
>At3g17450 unknown protein
Length = 877
Score = 311 bits (796), Expect = 1e-84
Identities = 185/559 (33%), Positives = 296/559 (52%), Gaps = 36/559 (6%)
Query: 39 DPAWEYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDV-----K 93
DP WE+ I+ + ++V+C +C +G R K HLA +VA C + P +V +
Sbjct: 133 DPGWEHGIAQDERKKKVKCNYCNKIVSGGINRFKQHLARIPGEVAPCKTAPEEVYVKIKE 192
Query: 94 KLMQDKVNKLQKKLLEKANLIVV-----DADGDKDGA------ADADLEIISGGR---GK 139
+ + K Q + ++ + D D ++D D ++ GR K
Sbjct: 193 NMKWHRAGKRQNRPDDEMGALTFRTVSQDPDQEEDREDHDFYPTSQDRLMLGNGRFSKDK 252
Query: 140 RKSAGEIAHKNMLKE-------------DSLR*NKAQSMVCGRRSTGKMLVRLLLSFFYN 186
RKS +++ + S + K S R + K + + F ++
Sbjct: 253 RKSFDSTNMRSVSEAKTKRARMIPFQSPSSSKQRKLYSSCSNRVVSRKDVTSSISKFLHH 312
Query: 187 NAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEK 246
+ A S + KM EL+ +G GF S + L I L +R+ W
Sbjct: 313 VGVPTEAANSLYFQKMIELIGMYGEGFVVPSSQLFSGRLLQEEMSTIKSYLREYRSSWVV 372
Query: 247 CGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGE 306
GC IM D WT+ + +++FLV P+G +F SID +DI + A +FK +D +V++IGE
Sbjct: 373 TGCSIMADTWTNTEGKKMISFLVSCPRGVYFHSSIDATDIVEDALSLFKCLDKLVDDIGE 432
Query: 307 ENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGK 366
ENVVQV+T N A +++AG+LL EKR +LYWTPCA HC +L+LEDF K+ ++ + K +
Sbjct: 433 ENVVQVITQNTAIFRSAGKLLEEKRKNLYWTPCAIHCTELVLEDF-SKLEFVSECLEKAQ 491
Query: 367 TITTYIYGRTSLISLL-HKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKSNEW 425
IT +IY +T L++L+ ++FT +D +RPA+ A+ + TL L +H+ L LF+S+ W
Sbjct: 492 RITRFIYNQTWLLNLMKNEFTQGLDLLRPAVMRHASGFTTLQSLMDHKASLRGLFQSDGW 551
Query: 426 KTSK-LAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMV-DWEEKAAMGYIYEE 483
S+ AKS++G+ V+ +VLS VFWK V L+ P+++V+ M+ D ++ +M Y Y
Sbjct: 552 ILSQTAAKSEEGREVEKMVLSAVFWKKVQYVLKSVDPVMQVIHMINDGGDRLSMPYAYGY 611
Query: 484 MDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFTVDDE 543
M AK I++ S AR Y P W++I+ RW+ H PL+ A F NP +Y P+F E
Sbjct: 612 MCCAKMAIKSIHSDDARKYGPFWRVIEYRWNPLFHHPLYVAAYFFNPAYKYRPDFMAQSE 671
Query: 544 IVNGMYACLRRMVADAEKR 562
+V G+ C+ R+ D +R
Sbjct: 672 VVRGVNECIVRLEPDNTRR 690
Score = 55.1 bits (131), Expect = 1e-07
Identities = 23/69 (33%), Positives = 39/69 (56%)
Query: 39 DPAWEYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKKLMQD 98
DP WE+ ++ + ++V+C +C +G YR+K HLA S +V C P +V M++
Sbjct: 11 DPGWEHGVAQDQRKKKVKCNYCGKIVSGGIYRLKQHLARVSGEVTYCDKSPEEVCMRMKE 70
Query: 99 KVNKLQKKL 107
+ + KKL
Sbjct: 71 NLVRSTKKL 79
>At3g22220 hypothetical protein
Length = 759
Score = 254 bits (649), Expect = 1e-67
Identities = 162/559 (28%), Positives = 275/559 (48%), Gaps = 42/559 (7%)
Query: 37 KTDPAWEYAISTEPGSR-RVQCKFCQLAFTGSAY-RVKHHLAGTSRDVAVCPSVPLDVKK 94
K D AW++ + G R +++C +C+ F G RVK HLAG +C VP +V+
Sbjct: 14 KQDSAWKHCEVYKYGDRVQMRCLYCRKMFKGGGITRVKEHLAGKKGQGTICDQVPDEVRL 73
Query: 95 LMQDKVN---KLQKKL---------------LEKANLIVVDADGDKDGAADADLEIISGG 136
+Q ++ + Q+K E + +D + + + ++
Sbjct: 74 FLQQCIDGTVRRQRKRRKSSPEPLPIAYFPPCEVETQVAASSDVNNGFKSPSSDVVVGQS 133
Query: 137 RGKRKSAGEIAHKNMLKEDS----------------LR*NKAQSMVCGRRSTGKMLVRLL 180
G+ K + KN E + + + +++V + V +
Sbjct: 134 TGRTKQRTYRSRKNNAFERNDLANVEVDRDMDNLIPVAISSVKNIVHPTSKEREKTVHMA 193
Query: 181 LS-FFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEA 239
+ F ++ F+ A S + + G G +++++R L E+ K+++
Sbjct: 194 MGRFLFDIGADFDAANSVNVQPFIDAIVSGGFGVSIPTHEDLRGWILKSCVEEVKKEIDE 253
Query: 240 HRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDD 299
+ +W++ GC ++ IL FLV+ P+ FLKS+D S+I + DK+++++ +
Sbjct: 254 CKTLWKRTGCSVLVQELNSNEGPLILKFLVYCPEKVVFLKSVDASEILDSEDKLYELLKE 313
Query: 300 VVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHT 359
VVEEIG+ NVVQV+T +Y AAG+ LM+ P LYW PCAAHCID MLE+F K+
Sbjct: 314 VVEEIGDTNVVQVITKCEDHYAAAGKKLMDVYPSLYWVPCAAHCIDKMLEEF-GKMDWIR 372
Query: 360 DAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDL 419
+ I + +T+T IY + +++L+ KFT D ++P T AT++ T+G + + + L +
Sbjct: 373 EIIEQARTVTRIIYNHSGVLNLMRKFTFGNDIVQPVCTSSATNFTTMGRIADLKPYLQAM 432
Query: 420 FKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKAAMGY 479
S+EW +K G + + + FWK + P+++VL +V E K AMGY
Sbjct: 433 VTSSEWNDCSYSKEAGGLAMTETINDEDFWKALTLANHITAPILRVLRIVCSERKPAMGY 492
Query: 480 IYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFT 539
+Y M RAKE I+TN + Y WKIID W L +PL+AAG +LNP Y+ +
Sbjct: 493 VYAAMYRAKEAIKTNLA-HREEYIVYWKIIDRWW---LQQPLYAAGFYLNPKFFYSIDEE 548
Query: 540 VDDEIVNGMYACLRRMVAD 558
+ EI + C+ ++V D
Sbjct: 549 MRSEIHLAVVDCIEKLVPD 567
>At4g15020 putative protein
Length = 522
Score = 202 bits (513), Expect = 6e-52
Identities = 152/542 (28%), Positives = 249/542 (45%), Gaps = 56/542 (10%)
Query: 26 ARVRPKNAKGNKTDPAWEYAISTEPGSR-RVQCKFCQLAFTGSAYRVKHHLAGTSRDVAV 84
A + P K D AW++ + G R +++C +C+ F G + RD
Sbjct: 3 AELEPVALTPQKQDNAWKHCEIYKYGDRLQMRCLYCRKMFKGGGITRVKDILLVKRDKQC 62
Query: 85 CPSVPLDVKKLMQDKVNKLQKKLLEKANLIVVDAD----------GDKD-GAADADLEII 133
+D Q K +K + L A+L ++ D G K G++D ++
Sbjct: 63 -----IDGTVRRQRKRHKSSSEPLSVASLPPIEGDMMVVQPDVNDGFKSPGSSDVVVQNE 117
Query: 134 SGGRGKRKSAGEIAHKNMLKEDSLR*N------KAQSMVCGRRSTGKMLVR--------- 178
S G+ K + KN + S N +++ S+ K +V
Sbjct: 118 SLLSGRTKQRTYRSKKNAFENGSASNNVDLIGRDMDNLIPVAISSVKNIVHPSFRDRENT 177
Query: 179 ---LLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGK 235
+ F + F+ S + M + +A G G ++D++R L E+ K
Sbjct: 178 IHMAIGRFLFGIGADFDAVNSVNFQPMIDAIASGGFGVSAPTHDDLRGWILKNCVEEMAK 237
Query: 236 DLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFK 295
+++ +A+W++ GC I+ + + +LNFLV+ P+ FLKS+D S++ +ADK+F+
Sbjct: 238 EIDECKAMWKRTGCSILVEELNSDKGFKVLNFLVYCPEKVVFLKSVDASEVLSSADKLFE 297
Query: 296 MIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKI 355
++ ++VEE+G NVVQV+T Y AG+ LM P LYW PCAAHCID MLE+F K+
Sbjct: 298 LLSELVEEVGSTNVVQVITKCDDYYVDAGKRLMLVYPSLYWVPCAAHCIDQMLEEF-GKL 356
Query: 356 PLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQ 415
++ I + + IT ++Y + A + AT++ TLG + E ++
Sbjct: 357 GWISETIEQAQAITRFVYNHS------------------AFSSSATNFATLGRIAELKSN 398
Query: 416 LIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKA 475
L + S EW ++ G +V N + + FWK V PL++ L +V E++
Sbjct: 399 LQAMVTSAEWNECSYSEEPSG-LVMNALTDEAFWKAVALVNHLTSPLLRALRIVCSEKRP 457
Query: 476 AMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYA 535
AMGY+Y + RAK+ I+T+ + Y WKIID G FLNP L Y
Sbjct: 458 AMGYVYAALYRAKDAIKTHLVNR-EDYIIYWKIIDHGGSNNSIFLCLLLGFFLNPKLFYN 516
Query: 536 PN 537
N
Sbjct: 517 TN 518
>At1g79740 hypothetical protein
Length = 518
Score = 191 bits (485), Expect = 1e-48
Identities = 108/362 (29%), Positives = 187/362 (50%), Gaps = 7/362 (1%)
Query: 202 MFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRR 261
M + VAK G GF S + ++L+ +I L+ W GC I+ + WTD +
Sbjct: 1 MLDAVAKCGPGFVAPSP---KTEWLDRVKSDISLQLKDTEKEWVTTGCTIIAEAWTDNKS 57
Query: 262 RTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYK 321
R ++NF V SP FF KS+D S K + + + D V+++IG+E++VQ++ DN+ Y
Sbjct: 58 RALINFSVSSPSRIFFHKSVDASSYFKNSKCLADLFDSVIQDIGQEHIVQIIMDNSFCYT 117
Query: 322 AAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISL 381
L++ ++ +PCA+ C++++LE+F K+ IS+ + I+ ++Y + ++ L
Sbjct: 118 GISNHLLQNYATIFVSPCASQCLNIILEEF-SKVDWVNQCISQAQVISKFVYNNSPVLDL 176
Query: 382 LHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQN 441
L K T D IR +T +++L+L + + + +L +F E+ T+ N
Sbjct: 177 LRKLTGGQDIIRSGVTRSVSNFLSLQSMMKQKARLKHMFNCPEYTTN--TNKPQSISCVN 234
Query: 442 VVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARS 501
++ FW+ V + + P++KVL V K A+G IYE M +AKE IRT +
Sbjct: 235 ILEDNDFWRAVEESVAISEPILKVLREVS-TGKPAVGSIYELMSKAKESIRTYYIMDENK 293
Query: 502 YNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEK 561
+ I+D W LH PLHAA FLNP ++Y P + + L +++ ++
Sbjct: 294 HKVFSDIVDTNWCEHLHSPLHAAAAFLNPSIQYNPEIKFLTSLKEDFFKVLEKLLPTSDL 353
Query: 562 RK 563
R+
Sbjct: 354 RR 355
>At3g13020 hypothetical protein
Length = 605
Score = 190 bits (482), Expect = 3e-48
Identities = 139/536 (25%), Positives = 249/536 (45%), Gaps = 45/536 (8%)
Query: 43 EYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKK-----LMQ 97
E+ I + RV+C +C S +R+KHHL DV C V L +++ LM+
Sbjct: 10 EHGICVDKKKSRVKCNYCGKEMN-SFHRLKHHLGAVGTDVTHCDQVSLTLRETFRTMLME 68
Query: 98 DKVNKLQKKLLEKANLIVVDADGDKDGAADADLEIISGGRGKRKSAGEIAHKNMLKEDSL 157
DK K + D+ + D+ + +S +G A E+ ++++L
Sbjct: 69 DKSGYTTPKTKRVGKFQMADSRKRRK-TEDSSSKSVSPEQGN--VAVEVDNQDLLS---- 121
Query: 158 R*NKAQSMVCGRRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLS 217
+KAQ + GR FFY + + + S + +M + G+G K
Sbjct: 122 --SKAQKCI-GR-------------FFYEHCVDLSAVDSPCFKEMMMAL---GVGQKIPD 162
Query: 218 YDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFF 277
++ + L E+ ++ + W+ GC I+ D W D + +++F+ P G +
Sbjct: 163 SHDLNGRLLQEAMKEVQDYVKNIKDSWKITGCSILLDAWIDPKGHDLVSFVADCPAGPVY 222
Query: 278 LKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAA-GELLMEKRPHLYW 336
LKSID S + + +++ +VEE+G NV Q++ + + + G+L ++W
Sbjct: 223 LKSIDVSVVKNDVTALLSLVNGLVEEVGVHNVTQIIACSTSGWVGELGKLFSGHDREVFW 282
Query: 337 TPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVD-SIRPA 395
+ +HC +LML K + D + K TI +I S + + ++ D ++ +
Sbjct: 283 SVSLSHCFELMLVKIGK-MRSFGDILDKVNTIWEFINNNPSALKIYRDQSHGKDITVSSS 341
Query: 396 MTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNC 455
F YL L + + + L +F S+ WK ++GK V N+V FW+ V
Sbjct: 342 EFEFVKPYLILKSVFKAKKNLAAMFASSVWK------KEEGKSVSNLVNDSSFWEAVEEI 395
Query: 456 LRGAFPLVKVLCMV-DWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWD 514
L+ PL L + + + +GYIY+ +D K I+ F+ + + Y +W +IDD W+
Sbjct: 396 LKCTSPLTDGLRLFSNADNNQHVGYIYDTLDGIKLSIKKEFNDEKKHYLTLWDVIDDVWN 455
Query: 515 RQLHRPLHAAGLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEKRKKKLISNL 570
+ LH PLHAAG +LNP Y+ +F +D E+ +G+ +V A++ + K+ S L
Sbjct: 456 KHLHNPLHAAGYYLNPTSFYSTDFHLDPEVSSGL---THSLVHVAKEGQIKIASQL 508
>At3g13030 unknown protein (At3g13030)
Length = 544
Score = 184 bits (467), Expect = 1e-46
Identities = 100/335 (29%), Positives = 165/335 (48%), Gaps = 9/335 (2%)
Query: 232 EIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTAD 291
E+ +E + W GC I+ D W D++ R ++ F+ P G +L S D SD
Sbjct: 104 EVQDRVEKIKESWAITGCSILLDAWVDQKGRDLVTFVADCPAGLVYLISFDVSDFKDDVT 163
Query: 292 KIFKMIDDVVEEIGEENVVQVVTDNAANYKAA-GELLMEKRPHLYWTPCAAHCIDLMLED 350
+ +++ +VEE+G NV Q++ + + + GEL ++W+ +HC +LML
Sbjct: 164 ALLSLVNGLVEEVGVRNVTQIIACSTSGWVGELGELFAGHDREVFWSVSVSHCFELMLVK 223
Query: 351 FEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVD-SIRPAMTHFATSYLTLGCL 409
K I D K I +I S++++ + +D ++ + F T YL L +
Sbjct: 224 ISK-IRSFGDIFDKVNNIWLFINNNPSVLNIFRDQCHGIDITVSSSEFEFVTPYLILESI 282
Query: 410 NEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMV 469
+ + L +F S+ W + + N+V FW+ V + L+ PL+ L +
Sbjct: 283 FKAKKNLTAMFASSNWNNEQCIA------ISNLVSDSSFWETVESVLKCTSPLIHGLLLF 336
Query: 470 DWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLN 529
+GY+Y+ MD KE I F+ + + Y P+W +IDD W++ LH PLHAAG FLN
Sbjct: 337 STANNQHLGYVYDTMDSIKESIAREFNHKPQFYKPLWDVIDDVWNKHLHNPLHAAGYFLN 396
Query: 530 PMLRYAPNFTVDDEIVNGMYACLRRMVADAEKRKK 564
P Y+ NF +D E+V G+ + L MV D + K
Sbjct: 397 PTAFYSTNFHLDIEVVTGLISSLIHMVEDCHVQFK 431
>At1g43260 hypothetical protein
Length = 294
Score = 153 bits (386), Expect = 3e-37
Identities = 89/273 (32%), Positives = 144/273 (52%), Gaps = 12/273 (4%)
Query: 168 GRRSTGKMLVRL------LLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEI 221
G + +ML RL + + Y++ I FN +D+ +M E+ + G G P S ++
Sbjct: 21 GANKSARMLERLHVVHQYVARWVYSHGIPFNAIANDDLRRMLEVAGQFGPGVTPPSQYQL 80
Query: 222 RNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSI 281
R L + +E W GC + TD W+DR+RR+I+N ++ +GT FL S
Sbjct: 81 REPLLKEEVVRMKGLMEEQEDEWRVNGCSVTTDSWSDRKRRSIMNLCINCKEGTMFLSSK 140
Query: 282 DPSDITKTADKIFKMIDDV-VEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCA 340
D D + T + IF +++ ++ +G ++VVQVVT+NA N A +LL E RP ++WT CA
Sbjct: 141 DCFDDSHTGEYIFAYVNEYCIKNLGGDHVVQVVTNNATNNITAAKLLKEVRPTIFWTFCA 200
Query: 341 AHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFA 400
H I+LM+E K+ + + + K T +IY +S++ FT D +RP + FA
Sbjct: 201 THTINLMVEGI-SKLAMSDEIVKMAKAFTIFIYAHHQTLSMMRSFTKRRDIVRPGIIGFA 259
Query: 401 TSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKS 433
+++ TL L E + L K K KL+K+
Sbjct: 260 SAFRTLKSLVEKEENL----KVKRKKKKKLSKA 288
>At3g13010 hypothetical protein
Length = 572
Score = 150 bits (379), Expect = 2e-36
Identities = 131/527 (24%), Positives = 219/527 (40%), Gaps = 71/527 (13%)
Query: 43 EYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKKLMQDKVNK 102
+Y E G+R VQC FC R+K+HL +DV C VPL +++ V
Sbjct: 10 DYGTWVEDGAR-VQCTFCDKRMN-DFNRLKYHLGNVCKDVTPCSQVPLPIREAFFKMV-- 65
Query: 103 LQKKLLEKANLIVVDADGDKDGAADADLEIISGGRGKRKSAGEIAHKNMLKEDSLR*NKA 162
++++ ++ RG R + G A + + + L A
Sbjct: 66 MEQRYPDRK-------------------------RG-RSNKGFTASETAAEAEKL----A 95
Query: 163 QSMVCGRRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIR 222
Q V FFY + + F+ S + KM + G G + D
Sbjct: 96 QMDVA--------------QFFYEHGVDFSAVDSTSFKKMMMIKTVGGEGGGQMIPD--- 138
Query: 223 NKYLN-YFYGEIGKDLEAH----RAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFF 277
++ LN + + E K ++ +A WE GC I+ D W + R ++ F+ P G +
Sbjct: 139 SRDLNGWMFQEALKKVQDRVKEIKASWEITGCSILFDAWIGPKGRDLVTFVADCPAGAVY 198
Query: 278 LKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWT 337
LKS D SDI + +++ +VEE+G NV Q++ + + + G+L + ++W+
Sbjct: 199 LKSADVSDIKTDVTALTSLVNGIVEEVGVRNVTQIIACSTSGW--VGDLGKQLAGQVFWS 256
Query: 338 PCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMT 397
++C+ LML + K D K K + I S + + + ++ VD +
Sbjct: 257 VSLSYCLKLMLVEIGKMYSFE-DIFEKVKLLLDLINNNPSFLYVFRENSHKVDV--SSEC 313
Query: 398 HFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLR 457
F YLTL + + + LF S EW K + G + + V FW+ + +
Sbjct: 314 EFVMPYLTLEHI--YWVRRAGLFASPEW------KKEQGIAISSFVNDSTFWESLDKIVG 365
Query: 458 GAFPLVKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQL 517
LV K + Y Y ++ K+ + F + + Y P W +IDD W
Sbjct: 366 STSSLVHGWLWFSRGSK-HVAYAYHFIESIKKNVAWTFKYERQFYEPTWNVIDDVWHNN- 423
Query: 518 HRPLHAAGLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEKRKK 564
H PLHAAG FLNPM Y+ +F + G+ L +V + + K
Sbjct: 424 HNPLHAAGYFLNPMAYYSDDFHTYQHVYTGLAFSLVHLVKEPHLQVK 470
>At3g48770 hypothetical protein
Length = 1899
Score = 43.9 bits (102), Expect = 3e-04
Identities = 35/111 (31%), Positives = 51/111 (45%), Gaps = 9/111 (8%)
Query: 36 NKTDPAWEYAISTEPGSRR--VQCKFCQLAFTGSAY-RVKHHLAGTSRDVAVCPSVPLDV 92
+K D AW Y RR + C FC + GS R+KHHLAG + A C V +V
Sbjct: 24 HKQDLAWNYVREAIDSRRRKVITCGFCGKQYVGSEINRMKHHLAGLNNRSA-CQKVSSEV 82
Query: 93 KKLMQDKVNKLQKKLLEKANLIVVDAD--GDKDGAADADLEIISGGRGKRK 141
+ M+ + + ++K +IVVD D D+ + + S KRK
Sbjct: 83 QHSMRKSLEENKEK---PKKIIVVDLDMQDDEVNVGQSSHQSCSSSSRKRK 130
>At4g05510
Length = 604
Score = 39.3 bits (90), Expect = 0.007
Identities = 60/274 (21%), Positives = 113/274 (40%), Gaps = 49/274 (17%)
Query: 201 KMFELVAKHGIGFKPLSYDEIRN--KYLNYFYGEIGKDLEAHRAV--WEKCGCIIMT--- 253
K ++ +H + F + Y+E+R+ Y+N Y ++ A V WEK I+ +
Sbjct: 103 KFSRVIIRHDLPFLCVEYEELRDFISYMNPDYKCYTRNTAAADVVKTWEKEKQILKSELE 162
Query: 254 ----------DGWTDRRRRTILNFLVHSPKGTFFLKS--IDPSDITK--TADKIFKMIDD 299
D WT + H + L S + SD+ T D + I +
Sbjct: 163 RIPSRICLTSDCWTSLGGDGYIVLTAHYVDTRWILNSKILSFSDMLPPHTGDALASKIHE 222
Query: 300 VVEEIG-EENVVQVVTDNAANYKAAGELLMEK---------RPHLYWTPCAAHCIDLMLE 349
++E G E+ V + DNA + E+L+++ + + C AH ++ +++
Sbjct: 223 CLKEWGIEKKVFTLTLDNATANNSMQEVLIDRLKLDNNLMCKGEFFHVRCCAHVLNRIVQ 282
Query: 350 DFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAM---THFATSYLTL 406
+ + + +DA+SK + Y+ G TS L + + ++ T + ++YL L
Sbjct: 283 N---GLDVISDALSKIRETVKYVKGSTSRRLALAECVEGKGEVLLSLDVQTRWNSTYLML 339
Query: 407 GCLNEHQNQLIDLFK-----------SNEWKTSK 429
++Q L + FK S EWK +K
Sbjct: 340 HKALKYQRAL-NRFKIVDKNYKNCPSSEEWKRAK 372
>At2g11910 unknown protein
Length = 168
Score = 35.4 bits (80), Expect = 0.11
Identities = 15/25 (60%), Positives = 21/25 (84%), Gaps = 1/25 (4%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQE 663
DEEGDNDD++ E+NEDE +DD ++
Sbjct: 132 DEEGDNDDED-EDNEDEEEDDDEED 155
>At5g40340 unknown protein
Length = 1008
Score = 33.5 bits (75), Expect = 0.40
Identities = 14/39 (35%), Positives = 25/39 (63%)
Query: 625 VNGIGVLLKGLPSIDEEGDNDDDEVEENEDEYDDDPYQE 663
+NG+ LLK ++EE + +++E EE ED D++ +E
Sbjct: 75 MNGVSSLLKLKEDVEEEEEEEEEEEEEEEDGEDEEEEEE 113
Score = 29.3 bits (64), Expect = 7.5
Identities = 15/64 (23%), Positives = 35/64 (54%)
Query: 600 LMDLDIRNCSGLL*EYLV*HVAHQGVNGIGVLLKGLPSIDEEGDNDDDEVEENEDEYDDD 659
++D+D +N L ++ GV+ + L + + +EE + +++E E+ EDE +++
Sbjct: 53 VVDVDEKNEKNLNESGVIEDCVMNGVSSLLKLKEDVEEEEEEEEEEEEEEEDGEDEEEEE 112
Query: 660 PYQE 663
+E
Sbjct: 113 EEEE 116
>At1g47970 Unknown protein (T2J15.12)
Length = 198
Score = 33.1 bits (74), Expect = 0.52
Identities = 13/19 (68%), Positives = 16/19 (83%)
Query: 641 EGDNDDDEVEENEDEYDDD 659
EGD+DDDE EE +D+ DDD
Sbjct: 48 EGDDDDDEEEEEDDDDDDD 66
Score = 29.6 bits (65), Expect = 5.8
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQE 663
DE+G+ DDD+ + ++D+ DDD E
Sbjct: 89 DEDGNGDDDDDDGDDDDDDDDDEDE 113
>At2g19480 putative nucleosome assembly protein
Length = 379
Score = 32.7 bits (73), Expect = 0.68
Identities = 13/25 (52%), Positives = 18/25 (72%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQE 663
D+E D +DDE +E ED+ DDD +E
Sbjct: 317 DDEEDEEDDEDDEEEDDEDDDEEEE 341
Score = 29.3 bits (64), Expect = 7.5
Identities = 11/26 (42%), Positives = 19/26 (72%)
Query: 638 IDEEGDNDDDEVEENEDEYDDDPYQE 663
IDE+ D +D+E +E+++E DD+ E
Sbjct: 313 IDEDDDEEDEEDDEDDEEEDDEDDDE 338
Score = 29.3 bits (64), Expect = 7.5
Identities = 12/25 (48%), Positives = 17/25 (68%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQE 663
D+E D DDDE +E +DE D++ E
Sbjct: 310 DDEIDEDDDEEDEEDDEDDEEEDDE 334
>At5g59000 unknown protein
Length = 231
Score = 32.3 bits (72), Expect = 0.89
Identities = 12/26 (46%), Positives = 21/26 (80%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQEF 664
+EE D+DDDE EE+E+E +++ +E+
Sbjct: 100 EEEDDSDDDEDEEDEEEEEEEEEEEY 125
Score = 30.0 bits (66), Expect = 4.4
Identities = 11/31 (35%), Positives = 20/31 (64%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQEFPPAMG 669
D+ D++D+E EE E+E +++ Y P +G
Sbjct: 103 DDSDDDEDEEDEEEEEEEEEEEYYGLPLQLG 133
>At4g04407 putative protein
Length = 298
Score = 32.3 bits (72), Expect = 0.89
Identities = 19/64 (29%), Positives = 28/64 (43%), Gaps = 14/64 (21%)
Query: 626 NGIGVLLKGLPSIDEEGDNDDD--------------EVEENEDEYDDDPYQEFPPAMGIN 671
+G V+ G D EGD+DDD E+EE++D D PY + N
Sbjct: 169 SGACVITNGGHDSDSEGDDDDDIPNAGIQNQGIEICEIEEDDDLQDYQPYSMYQGESDTN 228
Query: 672 DVLN 675
++ N
Sbjct: 229 ELFN 232
>At2g21420 Mutator-like transposase
Length = 468
Score = 32.3 bits (72), Expect = 0.89
Identities = 14/38 (36%), Positives = 23/38 (59%)
Query: 626 NGIGVLLKGLPSIDEEGDNDDDEVEENEDEYDDDPYQE 663
N G L G D++ D+DDD+ ++++D+ DDD E
Sbjct: 381 NQRGCLSPGCDDDDDDDDDDDDDDDDDDDDDDDDDDDE 418
Score = 29.3 bits (64), Expect = 7.5
Identities = 10/25 (40%), Positives = 19/25 (76%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQE 663
D++ D+DDD+ ++++D+ DDD E
Sbjct: 397 DDDDDDDDDDDDDDDDDDDDDEDDE 421
>At5g42280 putative protein
Length = 694
Score = 32.0 bits (71), Expect = 1.2
Identities = 11/21 (52%), Positives = 18/21 (85%)
Query: 639 DEEGDNDDDEVEENEDEYDDD 659
D++ DNDDD V++++D+ DDD
Sbjct: 170 DDDDDNDDDVVDDDDDDDDDD 190
>At5g17910 putative protein
Length = 1342
Score = 32.0 bits (71), Expect = 1.2
Identities = 12/24 (50%), Positives = 20/24 (83%)
Query: 640 EEGDNDDDEVEENEDEYDDDPYQE 663
EEGDN++++ EE+E+E D++ QE
Sbjct: 319 EEGDNENEDEEEDEEEEDEEEKQE 342
>At4g26600 unknown protein
Length = 671
Score = 32.0 bits (71), Expect = 1.2
Identities = 13/29 (44%), Positives = 20/29 (68%)
Query: 639 DEEGDNDDDEVEENEDEYDDDPYQEFPPA 667
DEEG+ND D+ ++++D+ DDD P A
Sbjct: 86 DEEGNNDSDDDDDDDDDDDDDDEDAEPLA 114
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,448,930
Number of Sequences: 26719
Number of extensions: 696004
Number of successful extensions: 4721
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 3924
Number of HSP's gapped (non-prelim): 486
length of query: 677
length of database: 11,318,596
effective HSP length: 106
effective length of query: 571
effective length of database: 8,486,382
effective search space: 4845724122
effective search space used: 4845724122
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC138452.7


