

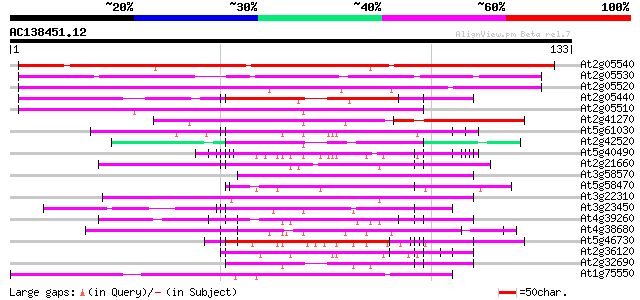
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138451.12 - phase: 0
(133 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g05540 putative glycine-rich protein 127 1e-30
At2g05530 putative glycine-rich protein 96 5e-21
At2g05520 putative glycine-rich protein 96 5e-21
At2g05440 putative glycine-rich protein 62 1e-10
At2g05510 putative glycine-rich protein 60 3e-10
At2g41270 pseudogene 57 2e-09
At5g61030 RNA-binding protein - like 56 6e-09
At2g42520 putative ATP-dependent RNA helicase 55 1e-08
At5g40490 ribonucleoprotein -like 52 7e-08
At2g21660 glycine-rich RNA binding protein 52 1e-07
At3g58570 ATP-dependent RNA helicase-like protein 51 2e-07
At5g58470 RNA/ssDNA-binding protein - like 50 3e-07
At3g22310 putative RNA helicase 50 3e-07
At3g23450 hypothetical protein 50 4e-07
At4g39260 glycine-rich protein (clone AtGRP8) 49 6e-07
At4g38680 glycine-rich protein 2 (GRP2) 49 6e-07
At5g46730 unknown protein 49 8e-07
At2g36120 unknown protein 49 1e-06
At2g32690 unknown protein 48 2e-06
At1g75550 hypothetical protein 48 2e-06
>At2g05540 putative glycine-rich protein
Length = 135
Score = 127 bits (320), Expect = 1e-30
Identities = 68/132 (51%), Positives = 85/132 (63%), Gaps = 8/132 (6%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASS---NTQKEADDQTNELNDAKYGSYGGGYP 59
+ KA+L L L+ ++LLI+SEV ARDL E S+ N +++ QT + G YGG +P
Sbjct: 2 ASKALLFLSLI-VVLLIASEVVARDLAEKSAEQKNNERDEVKQTEQFGGFPGGGYGG-FP 59
Query: 60 GRGGGGNYGGGYPRNRGGYPGNRGG--NYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCS 117
G G GGN GGGY GGY GG N GGGY GNRGG YCRYGCC YYG C +CC+
Sbjct: 60 GGGYGGNPGGGYGNRGGGYRNRDGGYGNRGGGY-GNRGGGYCRYGCCYRGYYGGCFRCCA 118
Query: 118 YAGEAVAMQTEN 129
YAG+AV Q ++
Sbjct: 119 YAGQAVQTQPQS 130
>At2g05530 putative glycine-rich protein
Length = 115
Score = 95.9 bits (237), Expect = 5e-21
Identities = 57/124 (45%), Positives = 72/124 (57%), Gaps = 13/124 (10%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRG 62
+ KA+++ GL A++L+++ EV+A T S + + DQ N G +GG G
Sbjct: 2 ASKALVLFGLFAVLLVVT-EVAAASGTVKSESGETVQPDQYN-------GGHGGN-GGYN 52
Query: 63 GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGEA 122
GGG Y GG N GGY N GG Y GG G R G YCRYGCC Y+G C +CCSYAGEA
Sbjct: 53 GGGGYNGGGGHNGGGY--NGGGGYNGGGHGGRHG-YCRYGCCYRGYHG-CSRCCSYAGEA 108
Query: 123 VAMQ 126
V Q
Sbjct: 109 VQTQ 112
>At2g05520 putative glycine-rich protein
Length = 145
Score = 95.9 bits (237), Expect = 5e-21
Identities = 57/142 (40%), Positives = 76/142 (53%), Gaps = 19/142 (13%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG-- 60
+ KA+++LGL A++L++S +A T S + + DQ ++ + GGGY G
Sbjct: 2 ASKALVLLGLFAVLLVVSEVAAASSATVNSESKETVKPDQRGYGDNGGNYNNGGGYQGGG 61
Query: 61 ----------RGGGGNYGGGYPRNRGG---YPGNRGGNYGGG---YPGNRGGNYCRYGCC 104
+GGGGNY GG R +GG Y G G GGG G GG+YCR+GCC
Sbjct: 62 GNYQGGGGNYQGGGGNYQGGGGRYQGGGGRYQGGGGRYQGGGGRQGGGGSGGSYCRHGCC 121
Query: 105 GDRYYGSCRKCCSYAGEAVAMQ 126
R Y C +CCSYAGEAV Q
Sbjct: 122 -YRGYNGCSRCCSYAGEAVQTQ 142
>At2g05440 putative glycine-rich protein
Length = 154
Score = 61.6 bits (148), Expect = 1e-10
Identities = 40/112 (35%), Positives = 58/112 (51%), Gaps = 12/112 (10%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRG 62
+ KA+++LGL +++L++S +AR S K ++T + YG GG+ G G
Sbjct: 2 ASKALILLGLFSVLLVVSEVSAARQ-----SGMVKPESEET--VQPEGYGGGHGGHGGHG 54
Query: 63 GGGNYGGGYPRNRGGYP----GNRGGNYGGGYPGNRGGNYCRYGCCGDRYYG 110
GGG +G G GG+ G GG+YGGG G+ GG YG G Y G
Sbjct: 55 GGGGHGHGGHNGGGGHGLDGYGGGGGHYGGG-GGHYGGGGGHYGGGGGHYGG 105
Score = 46.6 bits (109), Expect = 4e-06
Identities = 23/41 (56%), Positives = 26/41 (63%), Gaps = 5/41 (12%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPG 92
G YGGG G GGGG+YGGG GG G GG++GGG G
Sbjct: 108 GHYGGGGGGHGGGGHYGGG-----GGGYGGGGGHHGGGGHG 143
Score = 46.2 bits (108), Expect = 5e-06
Identities = 25/48 (52%), Positives = 28/48 (58%), Gaps = 6/48 (12%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
YG GG Y GGGG+YGGG G Y G GG+ GGG+ G GG Y
Sbjct: 89 YGGGGGHY--GGGGGHYGGG----GGHYGGGGGGHGGGGHYGGGGGGY 130
Score = 44.3 bits (103), Expect = 2e-05
Identities = 23/49 (46%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Query: 52 GSYGGGYPGRGGGGNY--GGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G YGGG GGGG + GGG GG+ G GG YGGG + GG +
Sbjct: 94 GHYGGGGGHYGGGGGHYGGGGGGHGGGGHYGGGGGGYGGGGGHHGGGGH 142
Score = 28.9 bits (63), Expect = 0.82
Identities = 14/24 (58%), Positives = 16/24 (66%), Gaps = 1/24 (4%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRN 74
YG GGGY G GGG + GGG+ N
Sbjct: 123 YGGGGGGYGG-GGGHHGGGGHGLN 145
>At2g05510 putative glycine-rich protein
Length = 127
Score = 60.5 bits (145), Expect = 3e-10
Identities = 36/103 (34%), Positives = 52/103 (49%), Gaps = 7/103 (6%)
Query: 3 SKKAILILGLLAIILLISSEVSARDL------TEASSNTQKEADDQTNELNDAKYGSYGG 56
+ KA+++LGL AI+L++S +AR +EA+ + YG G
Sbjct: 2 ASKALILLGLFAILLVVSEVSAARQSGMVKPESEATVQPEGYHGGHGGHGGGGHYGGGGH 61
Query: 57 GYPGRGGGGNYG-GGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G+ G GGG +G GY GG+ G GG+YGGG GG+Y
Sbjct: 62 GHGGHNGGGGHGLDGYGGGHGGHYGGGGGHYGGGGGHGGGGHY 104
>At2g41270 pseudogene
Length = 280
Score = 57.4 bits (137), Expect = 2e-09
Identities = 44/119 (36%), Positives = 51/119 (41%), Gaps = 35/119 (29%)
Query: 35 TQKEADD--QTNELNDAKYGSYGGGYPGRGGGGNYG------GGYPRNRGGY-------- 78
+Q EA + +T E ND + G G G GGGG G GG+ R R Y
Sbjct: 152 SQAEASEVVETVEPNDVEPQQGGRGGGGGGGGGRGGCRWGCCGGWWRGRCRYCCRSQAEA 211
Query: 79 ---------------PGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGEA 122
G RGG GGG G RGG CR+GCCG + G CR CC EA
Sbjct: 212 SEVVETVEPNDVEPQQGGRGGGGGGG--GGRGG--CRWGCCGGWWRGRCRYCCRSQAEA 266
Score = 57.4 bits (137), Expect = 2e-09
Identities = 44/119 (36%), Positives = 51/119 (41%), Gaps = 35/119 (29%)
Query: 35 TQKEADD--QTNELNDAKYGSYGGGYPGRGGGGNYG------GGYPRNRGGY-------- 78
+Q EA + +T E ND + G G G GGGG G GG+ R R Y
Sbjct: 97 SQAEASEVVETVEPNDVEPQQGGRGGGGGGGGGRGGCRWGCCGGWWRGRCRYCCRSQAEA 156
Query: 79 ---------------PGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGEA 122
G RGG GGG G RGG CR+GCCG + G CR CC EA
Sbjct: 157 SEVVETVEPNDVEPQQGGRGGGGGGG--GGRGG--CRWGCCGGWWRGRCRYCCRSQAEA 211
Score = 40.0 bits (92), Expect = 4e-04
Identities = 17/31 (54%), Positives = 19/31 (60%), Gaps = 2/31 (6%)
Query: 92 GNRGGNYCRYGCCGDRYYGSCRKCCSYAGEA 122
G RGG CR+GCCG + G CR CC EA
Sbjct: 73 GGRGG--CRWGCCGGWWRGRCRYCCRSQAEA 101
>At5g61030 RNA-binding protein - like
Length = 309
Score = 55.8 bits (133), Expect = 6e-09
Identities = 31/64 (48%), Positives = 33/64 (51%), Gaps = 4/64 (6%)
Query: 52 GSYGGGYPGRGGGGNYG---GGYPRNR-GGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDR 107
G YGGG G GG G YG GGY N GGY GN G YGG G GG+ +G G
Sbjct: 141 GGYGGGAGGYGGSGGYGGGAGGYGGNSGGGYGGNAAGGYGGSGAGGYGGDATGHGGAGGG 200
Query: 108 YYGS 111
Y S
Sbjct: 201 YGSS 204
Score = 54.3 bits (129), Expect = 2e-08
Identities = 37/92 (40%), Positives = 43/92 (46%), Gaps = 6/92 (6%)
Query: 20 SSEVSARDLTEASSNTQKE--ADDQTNE--LNDAKYGSYGGGYPGRGGGGNYGGGYPRNR 75
SS + A D + K A+D+T+ YG GGGY G GG G GGY +
Sbjct: 95 SSAIQALDGRDLHGRVVKVNYANDRTSGGGFGGGGYGGGGGGYGGSGGYGGGAGGYGGSG 154
Query: 76 G--GYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
G G G GGN GGGY GN G Y G G
Sbjct: 155 GYGGGAGGYGGNSGGGYGGNAAGGYGGSGAGG 186
Score = 48.1 bits (113), Expect = 1e-06
Identities = 30/72 (41%), Positives = 35/72 (47%), Gaps = 14/72 (19%)
Query: 51 YGSYGGGYPGRGG--------GGNYGGGYPRN-RGGYPGNRGGNYGGGYPGNRG-----G 96
YG GGY G GG GGN GGGY N GGY G+ G YGG G+ G G
Sbjct: 143 YGGGAGGYGGSGGYGGGAGGYGGNSGGGYGGNAAGGYGGSGAGGYGGDATGHGGAGGGYG 202
Query: 97 NYCRYGCCGDRY 108
+ +G G+ Y
Sbjct: 203 SSGGFGSSGNTY 214
Score = 34.3 bits (77), Expect = 0.019
Identities = 25/62 (40%), Positives = 28/62 (44%), Gaps = 5/62 (8%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNR--GGNYCRYGCCGDRYY 109
G+ GGY G G GG GG GG G G + G G GN G+ G GD Y
Sbjct: 173 GNAAGGYGGSGAGGY--GGDATGHGGAGGGYGSSGGFGSSGNTYGEGSSASAGAVGD-YN 229
Query: 110 GS 111
GS
Sbjct: 230 GS 231
Score = 30.0 bits (66), Expect = 0.37
Identities = 20/63 (31%), Positives = 26/63 (40%), Gaps = 5/63 (7%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRN---RGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDR 107
+G GGGY GG G+ G Y G G+ G+ G G G + G GD
Sbjct: 194 HGGAGGGYGSSGGFGSSGNTYGEGSSASAGAVGDYNGSSGYGSANTYGSS--NGGFAGDS 251
Query: 108 YYG 110
+G
Sbjct: 252 QFG 254
Score = 28.1 bits (61), Expect = 1.4
Identities = 22/66 (33%), Positives = 27/66 (40%), Gaps = 20/66 (30%)
Query: 45 ELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCC 104
++N A + GGG+ GGG YGGG GGGY G+ G G
Sbjct: 112 KVNYANDRTSGGGF----GGGGYGGG----------------GGGYGGSGGYGGGAGGYG 151
Query: 105 GDRYYG 110
G YG
Sbjct: 152 GSGGYG 157
>At2g42520 putative ATP-dependent RNA helicase
Length = 633
Score = 55.1 bits (131), Expect = 1e-08
Identities = 28/47 (59%), Positives = 28/47 (59%), Gaps = 7/47 (14%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G YGGG G GGGG YGGG GGY G GG YGGGY G G Y
Sbjct: 585 GGYGGGGGGYGGGGGYGGG-----GGYGG--GGGYGGGYGGASSGGY 624
Score = 43.5 bits (101), Expect = 3e-05
Identities = 36/104 (34%), Positives = 42/104 (39%), Gaps = 13/104 (12%)
Query: 25 ARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYG-------GGYPRNRGG 77
AR L E +E + A S+GGG R GG +G G + RGG
Sbjct: 530 ARPLAELMQEANQEVPEWLTRY--ASRSSFGGG-KNRRSGGRFGGRDFRREGSFGSGRGG 586
Query: 78 YPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGE 121
Y G GG YGGG GG Y G G Y G+ Y GE
Sbjct: 587 Y-GGGGGGYGGGGGYGGGGGYGGGGGYGGGYGGA--SSGGYGGE 627
Score = 32.0 bits (71), Expect = 0.097
Identities = 19/44 (43%), Positives = 21/44 (47%), Gaps = 6/44 (13%)
Query: 55 GGGYPGRGGGGNY--GGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
GG G GGGG Y G P + GY GG GGG+ GG
Sbjct: 68 GGSGVGVGGGGGYRADAGRPGSGSGY----GGRGGGGWNNRSGG 107
Score = 31.2 bits (69), Expect = 0.17
Identities = 17/30 (56%), Positives = 17/30 (56%), Gaps = 2/30 (6%)
Query: 51 YGSYGGGYPGRGG-GGNYGGGYPRNRGGYP 79
YG GGGY G GG GG YGG GG P
Sbjct: 600 YGG-GGGYGGGGGYGGGYGGASSGGYGGEP 628
Score = 28.9 bits (63), Expect = 0.82
Identities = 15/28 (53%), Positives = 16/28 (56%), Gaps = 2/28 (7%)
Query: 48 DAKYGSYGGGYPGRGGGG--NYGGGYPR 73
DA G GY GRGGGG N GG+ R
Sbjct: 83 DAGRPGSGSGYGGRGGGGWNNRSGGWDR 110
Score = 26.9 bits (58), Expect = 3.1
Identities = 13/32 (40%), Positives = 14/32 (43%)
Query: 55 GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNY 86
GGGY G G GY GG NR G +
Sbjct: 77 GGGYRADAGRPGSGSGYGGRGGGGWNNRSGGW 108
>At5g40490 ribonucleoprotein -like
Length = 423
Score = 52.4 bits (124), Expect = 7e-08
Identities = 33/73 (45%), Positives = 34/73 (46%), Gaps = 12/73 (16%)
Query: 48 DAKYGSYGGGYPGRGG----GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG-------- 95
D G +GGGY G GG GG YGGG GGY G GG GGGY G G
Sbjct: 234 DGYGGGHGGGYGGPGGPYKSGGGYGGGRSGGYGGYGGEFGGYGGGGYGGGVGPYRGEPAL 293
Query: 96 GNYCRYGCCGDRY 108
G RYG G Y
Sbjct: 294 GYSGRYGGGGGGY 306
Score = 50.1 bits (118), Expect = 3e-07
Identities = 34/69 (49%), Positives = 35/69 (50%), Gaps = 11/69 (15%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRG----GYPGNRGGNYG----GGYPGNRGGNYCRYG 102
YG YGG + G GGGG YGGG RG GY G GG G GGY GG Y G
Sbjct: 265 YGGYGGEFGGYGGGG-YGGGVGPYRGEPALGYSGRYGGGGGGYNRGGYSMGGGGGY--GG 321
Query: 103 CCGDRYYGS 111
GD Y GS
Sbjct: 322 GPGDMYGGS 330
Score = 48.5 bits (114), Expect = 1e-06
Identities = 29/68 (42%), Positives = 35/68 (50%), Gaps = 3/68 (4%)
Query: 45 ELNDAKYGSYGGGYPGRGGGGNYGGGYPRNR-GGYPGNRGGNYGGGYPGNRGGNYCRYGC 103
++ YG GGGY G G +YGGGY + GGY G GG GGGY G G + G
Sbjct: 325 DMYGGSYGEPGGGYGGPSG--SYGGGYGSSGIGGYGGGMGGAGGGGYRGGGGYDMGGVGG 382
Query: 104 CGDRYYGS 111
G YG+
Sbjct: 383 GGAGGYGA 390
Score = 48.5 bits (114), Expect = 1e-06
Identities = 29/67 (43%), Positives = 32/67 (47%), Gaps = 13/67 (19%)
Query: 52 GSYGGGYPGRG-----------GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCR 100
GSYGGGY G GGG Y GG + GG G G YG G GN GG++
Sbjct: 343 GSYGGGYGSSGIGGYGGGMGGAGGGGYRGGGGYDMGGVGGGGAGGYGAGGGGNGGGSF-- 400
Query: 101 YGCCGDR 107
YG G R
Sbjct: 401 YGGGGGR 407
Score = 47.4 bits (111), Expect = 2e-06
Identities = 27/58 (46%), Positives = 31/58 (52%), Gaps = 3/58 (5%)
Query: 53 SYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYG 110
++GGGY G G GG +GGGY G Y GG YGGG G GG +G G YG
Sbjct: 227 NFGGGY-GDGYGGGHGGGYGGPGGPYKS--GGGYGGGRSGGYGGYGGEFGGYGGGGYG 281
Score = 47.4 bits (111), Expect = 2e-06
Identities = 29/58 (50%), Positives = 30/58 (51%), Gaps = 15/58 (25%)
Query: 52 GSYGGG-----------YPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G YGGG Y GR GGG GGGY NRGGY GG YGGG GG+Y
Sbjct: 278 GGYGGGVGPYRGEPALGYSGRYGGG--GGGY--NRGGYSMGGGGGYGGGPGDMYGGSY 331
Score = 47.0 bits (110), Expect = 3e-06
Identities = 28/60 (46%), Positives = 30/60 (49%), Gaps = 5/60 (8%)
Query: 50 KYGSYGGGYP----GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
+YG GGGY GGGG YGGG GG G GG YGG G+ GG Y G G
Sbjct: 298 RYGGGGGGYNRGGYSMGGGGGYGGGPGDMYGGSYGEPGGGYGGP-SGSYGGGYGSSGIGG 356
Score = 46.2 bits (108), Expect = 5e-06
Identities = 25/46 (54%), Positives = 27/46 (58%), Gaps = 2/46 (4%)
Query: 52 GSYGGGYPGRGGGGNYGGGYP-RNRGGYPGNRGGNYGGGYPGNRGG 96
G YG GY G GGG G G P ++ GGY G R G Y GGY G GG
Sbjct: 230 GGYGDGYGGGHGGGYGGPGGPYKSGGGYGGGRSGGY-GGYGGEFGG 274
Score = 45.8 bits (107), Expect = 6e-06
Identities = 28/65 (43%), Positives = 30/65 (46%), Gaps = 7/65 (10%)
Query: 52 GSYGGGYPGRGGGG-NYGGGYPRN------RGGYPGNRGGNYGGGYPGNRGGNYCRYGCC 104
G YGGG G GGGG GGGY GGY GGN GG + G GG G
Sbjct: 355 GGYGGGMGGAGGGGYRGGGGYDMGGVGGGGAGGYGAGGGGNGGGSFYGGGGGRGGYGGGG 414
Query: 105 GDRYY 109
RY+
Sbjct: 415 SGRYH 419
Score = 44.3 bits (103), Expect = 2e-05
Identities = 27/54 (50%), Positives = 28/54 (51%), Gaps = 8/54 (14%)
Query: 52 GSYGGGYPGRGGGGNYG---GGYPRNRGGYPGNRG----GNYGGGYPGNRGGNY 98
G YGGG PG GG+YG GGY G Y G G G YGGG G GG Y
Sbjct: 317 GGYGGG-PGDMYGGSYGEPGGGYGGPSGSYGGGYGSSGIGGYGGGMGGAGGGGY 369
Score = 43.5 bits (101), Expect = 3e-05
Identities = 23/45 (51%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query: 54 YGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
Y G Y G GGG N GG GGY G G YGG Y G GG Y
Sbjct: 295 YSGRYGGGGGGYNRGGYSMGGGGGYGGGPGDMYGGSY-GEPGGGY 338
Score = 36.2 bits (82), Expect = 0.005
Identities = 20/43 (46%), Positives = 22/43 (50%), Gaps = 1/43 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNR 94
G GGY G GGGGN GG + GG G GG G +P R
Sbjct: 382 GGGAGGY-GAGGGGNGGGSFYGGGGGRGGYGGGGSGRYHPYGR 423
Score = 29.3 bits (64), Expect = 0.63
Identities = 12/23 (52%), Positives = 12/23 (52%)
Query: 76 GGYPGNRGGNYGGGYPGNRGGNY 98
G N GG YG GY G GG Y
Sbjct: 222 GDSRSNFGGGYGDGYGGGHGGGY 244
Score = 27.3 bits (59), Expect = 2.4
Identities = 19/56 (33%), Positives = 21/56 (36%), Gaps = 11/56 (19%)
Query: 66 NYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGE 121
N+GGGY G YGGG+ G GG Y G G Y GE
Sbjct: 227 NFGGGY-----------GDGYGGGHGGGYGGPGGPYKSGGGYGGGRSGGYGGYGGE 271
>At2g21660 glycine-rich RNA binding protein
Length = 176
Score = 51.6 bits (122), Expect = 1e-07
Identities = 26/47 (55%), Positives = 27/47 (57%), Gaps = 3/47 (6%)
Query: 51 YGSYGGGYPGRGGGG-NYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
Y GGGY RGGGG +YGGG GGY G GG YGG G GG
Sbjct: 131 YSGGGGGYSSRGGGGGSYGGGRREGGGGYGGGEGGGYGGS--GGGGG 175
Score = 49.3 bits (116), Expect = 6e-07
Identities = 29/53 (54%), Positives = 31/53 (57%), Gaps = 7/53 (13%)
Query: 52 GSYGGGYPGRGGGGNY---GGGYPRNRGGYPGNRGGNY---GGGYPGNRGGNY 98
GSYGGG R GGG Y GGGY +RGG G+ GG GGGY G GG Y
Sbjct: 116 GSYGGGGGRREGGGGYSGGGGGYS-SRGGGGGSYGGGRREGGGGYGGGEGGGY 167
Score = 47.0 bits (110), Expect = 3e-06
Identities = 31/96 (32%), Positives = 39/96 (40%), Gaps = 3/96 (3%)
Query: 22 EVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGG---NYGGGYPRNRGGY 78
E + +D E + + T ++ GGG+ G GGGG GGGY G Y
Sbjct: 59 EKAMKDAIEGMNGQDLDGRSITVNEAQSRGSGGGGGHRGGGGGGYRSGGGGGYSGGGGSY 118
Query: 79 PGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRK 114
G G GGG GG Y G G Y G R+
Sbjct: 119 GGGGGRREGGGGYSGGGGGYSSRGGGGGSYGGGRRE 154
Score = 26.9 bits (58), Expect = 3.1
Identities = 10/16 (62%), Positives = 11/16 (68%)
Query: 52 GSYGGGYPGRGGGGNY 67
G GGGY G GGGG +
Sbjct: 161 GGEGGGYGGSGGGGGW 176
Score = 26.6 bits (57), Expect = 4.1
Identities = 11/20 (55%), Positives = 12/20 (60%)
Query: 52 GSYGGGYPGRGGGGNYGGGY 71
G YGGG G GG GGG+
Sbjct: 157 GGYGGGEGGGYGGSGGGGGW 176
>At3g58570 ATP-dependent RNA helicase-like protein
Length = 646
Score = 51.2 bits (121), Expect = 2e-07
Identities = 27/56 (48%), Positives = 27/56 (48%)
Query: 55 GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYG 110
GGG GGGG YGG G PG G GGGY GG Y YG G YYG
Sbjct: 570 GGGADYYGGGGGYGGVPGGGYGAMPGGYGPVPGGGYGNVPGGGYAPYGRGGGAYYG 625
Score = 32.7 bits (73), Expect = 0.057
Identities = 23/45 (51%), Positives = 24/45 (53%), Gaps = 10/45 (22%)
Query: 51 YGSYGGGY---PGRGGGGNYGGGY-PRNRGGYPGNRGGNYG-GGY 90
YG+ GGY PG G G GGGY P RGG G YG GGY
Sbjct: 590 YGAMPGGYGPVPGGGYGNVPGGGYAPYGRGG-----GAYYGPGGY 629
Score = 31.6 bits (70), Expect = 0.13
Identities = 21/40 (52%), Positives = 21/40 (52%), Gaps = 10/40 (25%)
Query: 52 GSYGG---GYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGG 88
G YGG GY GRG G YGG RG G RGG GG
Sbjct: 56 GGYGGANSGYGGRGQG--YGG-----RGSGYGGRGGPVGG 88
Score = 29.6 bits (65), Expect = 0.48
Identities = 20/41 (48%), Positives = 20/41 (48%), Gaps = 9/41 (21%)
Query: 59 PGRGGGGNYGGGYPRNRGGYPGNRGGNYGG---GYPGNRGG 96
PG G YGG GY G RG YGG GY G RGG
Sbjct: 50 PGNNDRGGYGGA----NSGY-GGRGQGYGGRGSGY-GGRGG 84
Score = 29.3 bits (64), Expect = 0.63
Identities = 18/38 (47%), Positives = 18/38 (47%), Gaps = 7/38 (18%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGG 88
YG G GY GRG G YGG RGG G GG
Sbjct: 65 YGGRGQGYGGRGSG--YGG-----RGGPVGGWNARSGG 95
>At5g58470 RNA/ssDNA-binding protein - like
Length = 422
Score = 50.1 bits (118), Expect = 3e-07
Identities = 35/87 (40%), Positives = 40/87 (45%), Gaps = 19/87 (21%)
Query: 52 GSYGGG---YPGRG--GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGN--------- 97
G YGGG Y GRG GGG+YGG GG GNRGG GG G+RGG
Sbjct: 27 GGYGGGDAGYGGRGASGGGSYGGRGGYGGGGGRGNRGGGGGGYQGGDRGGRGSGGGGRDG 86
Query: 98 --YCRYGCCGDRYYG---SCRKCCSYA 119
C CG+ + C KC + A
Sbjct: 87 DWRCPNPSCGNVNFARRVECNKCGALA 113
Score = 45.4 bits (106), Expect = 8e-06
Identities = 27/50 (54%), Positives = 28/50 (56%), Gaps = 8/50 (16%)
Query: 53 SYGGGYPGRGGGGNYGGGYP------RNRGGYPGNRGGNYGGGYPGNRGG 96
SYGG G GGGG YGGG + GG G RGG GGG GNRGG
Sbjct: 17 SYGGD--GYGGGGGYGGGDAGYGGRGASGGGSYGGRGGYGGGGGRGNRGG 64
Score = 33.9 bits (76), Expect = 0.025
Identities = 20/47 (42%), Positives = 23/47 (48%), Gaps = 2/47 (4%)
Query: 53 SYGGGYPGRGG-GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
SY GG P GG G YG P + GG G GG GG P + +Y
Sbjct: 213 SYSGGPPSYGGPRGGYGSDAP-STGGRGGRSGGYDGGSAPRRQEASY 258
Score = 32.3 bits (72), Expect = 0.074
Identities = 16/34 (47%), Positives = 17/34 (49%)
Query: 56 GGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGG 89
GG G GGGG GGG R R Y N+ GG
Sbjct: 383 GGGRGGGGGGYGGGGGDRRRDNYSSGPDRNHHGG 416
Score = 31.2 bits (69), Expect = 0.17
Identities = 20/45 (44%), Positives = 21/45 (46%), Gaps = 8/45 (17%)
Query: 48 DAKYGSYGGGYPGRGGGG------NYGGGYPRNRGGYPGNRGGNY 86
D + G GGG G GGGG NY G RN G GNR Y
Sbjct: 380 DQRGGGRGGGGGGYGGGGGDRRRDNYSSGPDRNHHG--GNRSRPY 422
Score = 30.0 bits (66), Expect = 0.37
Identities = 20/54 (37%), Positives = 22/54 (40%), Gaps = 2/54 (3%)
Query: 60 GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRY--GCCGDRYYGS 111
G G N GG +RGG +RGG GG R RY G YGS
Sbjct: 116 GTSSGANDRGGGGYSRGGGDSDRGGGRGGRNDSGRSYESSRYDGGSRSGGSYGS 169
Score = 29.3 bits (64), Expect = 0.63
Identities = 24/62 (38%), Positives = 28/62 (44%), Gaps = 3/62 (4%)
Query: 57 GYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCC 116
G RGGGG GG +RGG G R + G Y +R R G G GS R+
Sbjct: 120 GANDRGGGGYSRGGGDSDRGGGRGGRNDS-GRSYESSRYDGGSRSG--GSYGSGSQRENG 176
Query: 117 SY 118
SY
Sbjct: 177 SY 178
Score = 29.3 bits (64), Expect = 0.63
Identities = 20/55 (36%), Positives = 23/55 (41%), Gaps = 6/55 (10%)
Query: 47 NDAKYGSY--GGGYPGRGGG----GNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG 95
ND G Y GGG RGGG + G Y +R GG+YG G G
Sbjct: 122 NDRGGGGYSRGGGDSDRGGGRGGRNDSGRSYESSRYDGGSRSGGSYGSGSQRENG 176
Score = 28.9 bits (63), Expect = 0.82
Identities = 23/56 (41%), Positives = 24/56 (42%), Gaps = 12/56 (21%)
Query: 56 GGYPGRGGGG----NYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDR 107
G Y GGGG +YGG GY G GG YGGG G G G G R
Sbjct: 3 GMYNQDGGGGAPIPSYGGD------GYGG--GGGYGGGDAGYGGRGASGGGSYGGR 50
Score = 28.5 bits (62), Expect = 1.1
Identities = 13/24 (54%), Positives = 13/24 (54%)
Query: 75 RGGYPGNRGGNYGGGYPGNRGGNY 98
RGG G GG YGGG R NY
Sbjct: 382 RGGGRGGGGGGYGGGGGDRRRDNY 405
Score = 26.6 bits (57), Expect = 4.1
Identities = 13/35 (37%), Positives = 16/35 (45%), Gaps = 2/35 (5%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNY 86
G YG P GG G GGY + G P + +Y
Sbjct: 226 GGYGSDAPSTGGRGGRSGGY--DGGSAPRRQEASY 258
Score = 25.8 bits (55), Expect = 6.9
Identities = 19/43 (44%), Positives = 19/43 (44%), Gaps = 10/43 (23%)
Query: 63 GGGNYGGGYPRNRGGYPGNRG----GNYGGGYPGN-RGGNYCR 100
GGG GGG GGY G G NY G N GGN R
Sbjct: 383 GGGRGGGG-----GGYGGGGGDRRRDNYSSGPDRNHHGGNRSR 420
>At3g22310 putative RNA helicase
Length = 610
Score = 50.1 bits (118), Expect = 3e-07
Identities = 36/109 (33%), Positives = 45/109 (41%), Gaps = 21/109 (19%)
Query: 23 VSARDLTEASSNTQKEADDQTNELNDAKY----------------GSYGGGYPGRGGGGN 66
+ +D T A +KE + NEL GS+GGG G GG G+
Sbjct: 463 IHGQDQTRAVKMIEKEVGSRFNELPSIAVERGSASMFEGVGARSGGSFGGGRSGGGGYGS 522
Query: 67 YGGGYPRNRGGYPGNRGGNY-----GGGYPGNRGGNYCRYGCCGDRYYG 110
YG R+ GG G GG+ GGG G GG+ RY DR G
Sbjct: 523 YGSSSGRSGGGSYGGYGGSSGRSGGGGGSYGGSGGSSSRYSGGSDRSSG 571
Score = 32.0 bits (71), Expect = 0.097
Identities = 21/51 (41%), Positives = 25/51 (48%), Gaps = 7/51 (13%)
Query: 51 YGSYGGGYPGRGGG----GNYGGGYPRNRGGYPGNRG-GNYGGGYPGNRGG 96
YG YGG GGG G GG R GG + G G++G G G+ GG
Sbjct: 535 YGGYGGSSGRSGGGGGSYGGSGGSSSRYSGGSDRSSGFGSFGSG--GSSGG 583
Score = 30.0 bits (66), Expect = 0.37
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Query: 47 NDAKYGSYGGGYPGRGGGGN-YGGGYPRNRGGYPGNRGGNYGG 88
+ + G GG Y G GG + Y GG R+ G GG+ GG
Sbjct: 541 SSGRSGGGGGSYGGSGGSSSRYSGGSDRSSGFGSFGSGGSSGG 583
>At3g23450 hypothetical protein
Length = 452
Score = 49.7 bits (117), Expect = 4e-07
Identities = 23/45 (51%), Positives = 25/45 (55%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G +GGG G GGGG GGG GG+ G GG GGG G GG
Sbjct: 74 GGFGGGGGGGGGGGGGGGGGFGGGGGFGGGHGGGVGGGVGGGHGG 118
Score = 47.8 bits (112), Expect = 2e-06
Identities = 31/88 (35%), Positives = 44/88 (49%), Gaps = 11/88 (12%)
Query: 9 ILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYG 68
+L LL + + + V ARD++ +D+ + K G +GGG+ G G GG G
Sbjct: 10 LLALLCFHVFVVNVV-ARDVSSGRD------EDEKTLVGGGKGGGFGGGFGG-GAGGGVG 61
Query: 69 GGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
GG GG+ G GG +GGG G GG
Sbjct: 62 GG---AGGGFGGGAGGGFGGGGGGGGGG 86
Score = 44.7 bits (104), Expect = 1e-05
Identities = 25/56 (44%), Positives = 30/56 (52%), Gaps = 1/56 (1%)
Query: 51 YGSYGGGYPGRGGGGNYG-GGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
+G GGG G GGGG G GG GG+ G GG GGG+ G GG + + G G
Sbjct: 76 FGGGGGGGGGGGGGGGGGFGGGGGFGGGHGGGVGGGVGGGHGGGVGGGFGKGGGIG 131
Score = 41.6 bits (96), Expect = 1e-04
Identities = 21/45 (46%), Positives = 23/45 (50%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG+ G GGG GGG GG G G GGG+ G GG
Sbjct: 62 GGAGGGFGGGAGGGFGGGGGGGGGGGGGGGGGFGGGGGFGGGHGG 106
Score = 40.0 bits (92), Expect = 4e-04
Identities = 23/46 (50%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query: 52 GSYGGGYPGR-GGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G +GGG G GGGG GGG GG G GG +GGG+ G GG
Sbjct: 66 GGFGGGAGGGFGGGGGGGGGGGGGGGGGFGG-GGGFGGGHGGGVGG 110
Score = 39.3 bits (90), Expect = 6e-04
Identities = 21/49 (42%), Positives = 27/49 (54%), Gaps = 3/49 (6%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPG---NRGGNYGGGYPGNRGG 96
+G GG G GGGG +GGG +GG G +GG +GGG +GG
Sbjct: 368 FGKGGGIGGGIGGGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGG 416
Score = 38.9 bits (89), Expect = 8e-04
Identities = 23/47 (48%), Positives = 26/47 (54%), Gaps = 3/47 (6%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
K G +GGG G G GG GGG +GG G GG +GGG G GG
Sbjct: 402 KGGGFGGG-GGFGKGGGIGGGGGFGKGG--GFGGGGFGGGGGGGGGG 445
Score = 38.5 bits (88), Expect = 0.001
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query: 60 GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G G GG +GGG+ GG G GG GGG+ G GG +
Sbjct: 41 GGGKGGGFGGGFG---GGAGGGVGGGAGGGFGGGAGGGF 76
Score = 37.7 bits (86), Expect = 0.002
Identities = 21/45 (46%), Positives = 25/45 (54%), Gaps = 1/45 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G+GGG GGG+ + GG+ G G GGG G GG
Sbjct: 405 GFGGGGGFGKGGGIGGGGGFGKG-GGFGGGGFGGGGGGGGGGGGG 448
Score = 36.6 bits (83), Expect = 0.004
Identities = 24/55 (43%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGG-GYPGNRGGNYCRYGCCG 105
G +GGG+ G G GG GGG+ GG G GG GG G G GG + G G
Sbjct: 98 GGFGGGHGG-GVGGGVGGGHGGGVGGGFGKGGGIGGGIGKGGGVGGGIGKGGGIG 151
Score = 36.6 bits (83), Expect = 0.004
Identities = 21/51 (41%), Positives = 26/51 (50%), Gaps = 5/51 (9%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPG-----NRGGNYGGGYPGNRGG 96
+G GG G G GG +GGG +GG G +GG +GGG G GG
Sbjct: 390 FGKGGGIGGGIGKGGGFGGGGGFGKGGGIGGGGGFGKGGGFGGGGFGGGGG 440
Score = 35.4 bits (80), Expect = 0.009
Identities = 23/49 (46%), Positives = 26/49 (52%), Gaps = 3/49 (6%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRG--GYPGNRGGNYGGGYPGNRGG 96
K G GGG G+GGG GGG+ + G G G GG GGG G GG
Sbjct: 306 KGGGIGGGI-GKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGGGFGKGGG 353
Score = 34.7 bits (78), Expect = 0.015
Identities = 23/55 (41%), Positives = 29/55 (51%), Gaps = 5/55 (9%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
+G GG G G GG GGG+ + GG G GG GGG+ G GG + + G G
Sbjct: 348 FGKGGGIGGGIGKGGGIGGGFGKG-GGIGGGIGG--GGGFGG--GGGFGKGGGIG 397
Score = 34.7 bits (78), Expect = 0.015
Identities = 23/48 (47%), Positives = 23/48 (47%), Gaps = 1/48 (2%)
Query: 50 KYGSYGGGYPGRGG-GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
K G GGG G GG GG G G GG G GG GGG G GG
Sbjct: 370 KGGGIGGGIGGGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGGG 417
Score = 33.5 bits (75), Expect = 0.033
Identities = 21/49 (42%), Positives = 25/49 (50%), Gaps = 4/49 (8%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPG---NRGGNYGGGYPGNRGG 96
+G GG G G GG GGG +GG G +GG GGG+ G GG
Sbjct: 326 FGKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGF-GKGGG 373
Score = 33.1 bits (74), Expect = 0.043
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Query: 52 GSYGGGYPGRGG-GGNYGGGYPRNRGGYPG---NRGGNYGGGYPGNRGG 96
G +GGG GG GG GGG GG G +GG GGG G GG
Sbjct: 92 GGFGGGGGFGGGHGGGVGGGVGGGHGGGVGGGFGKGGGIGGGI-GKGGG 139
Score = 33.1 bits (74), Expect = 0.043
Identities = 22/49 (44%), Positives = 26/49 (52%), Gaps = 7/49 (14%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
K G GGG G GG GGG+ + GG G +GG GGG+ G GG
Sbjct: 196 KGGGIGGGI---GKGGGVGGGFGKG-GGVGGGIGKGGGVGGGF-GKGGG 239
Score = 32.7 bits (73), Expect = 0.057
Identities = 22/47 (46%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
K G GGG G+GGG G G GG G GG GGG G GG
Sbjct: 286 KGGGIGGGI-GKGGGIGGGIGKGGGIGGGIGKGGGIGGGGGFGKGGG 331
Score = 32.7 bits (73), Expect = 0.057
Identities = 21/48 (43%), Positives = 25/48 (51%), Gaps = 4/48 (8%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
+G GG G G GG GGG+ + GG G +GG GGG G GG
Sbjct: 214 FGKGGGVGGGIGKGGGVGGGFGKG-GGVGGGIGKGGGIGGGI-GKGGG 259
Score = 32.3 bits (72), Expect = 0.074
Identities = 24/64 (37%), Positives = 29/64 (44%), Gaps = 8/64 (12%)
Query: 52 GSYGGGYPGRGG-----GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGD 106
G GGG GG GG GGG + GG+ G G GGG G GG + + G G
Sbjct: 376 GGIGGGGGFGGGGGFGKGGGIGGGIGKG-GGFGGGGGFGKGGGIGG--GGGFGKGGGFGG 432
Query: 107 RYYG 110
+G
Sbjct: 433 GGFG 436
Score = 32.3 bits (72), Expect = 0.074
Identities = 19/40 (47%), Positives = 21/40 (52%), Gaps = 7/40 (17%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGG 89
K G GGG G G GG +GG GG+ G GG GGG
Sbjct: 414 KGGGIGGG-GGFGKGGGFGG------GGFGGGGGGGGGGG 446
Score = 32.3 bits (72), Expect = 0.074
Identities = 22/47 (46%), Positives = 23/47 (48%), Gaps = 2/47 (4%)
Query: 52 GSYGGGYPGRGGG--GNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G+GGG G G G GG G GG GGG G GG
Sbjct: 383 GFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGGGIGGGGGFGKGGG 429
Score = 32.0 bits (71), Expect = 0.097
Identities = 22/48 (45%), Positives = 25/48 (51%), Gaps = 5/48 (10%)
Query: 52 GSYGGGYPGR-GGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
G +GGG G G GG GGG + GG G +GG GGG G GG
Sbjct: 114 GGHGGGVGGGFGKGGGIGGGIGKG-GGVGGGIGKGGGIGGGI-GKGGG 159
Score = 31.6 bits (70), Expect = 0.13
Identities = 22/49 (44%), Positives = 25/49 (50%), Gaps = 7/49 (14%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
K G GGG+ G GG GGG + GG G +GG GGG G GG
Sbjct: 226 KGGGVGGGF---GKGGGVGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 269
Score = 31.6 bits (70), Expect = 0.13
Identities = 21/48 (43%), Positives = 24/48 (49%), Gaps = 5/48 (10%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRG-GYPGNRGGNYGGGYPGNRGG 96
K G GGG+ G GG GGG + G G +GG GGG G GG
Sbjct: 206 KGGGVGGGF---GKGGGVGGGIGKGGGVGGGFGKGGGVGGGI-GKGGG 249
Score = 31.2 bits (69), Expect = 0.17
Identities = 24/60 (40%), Positives = 28/60 (46%), Gaps = 8/60 (13%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGY--PGNRGGNYCRYGCCG 105
K G GGG G GG GGG + GG G +GG GGG G GG + + G G
Sbjct: 166 KGGGIGGGI---GKGGGIGGGIGKG-GGIGGGIGKGGGIGGGIGKGGGVGGGFGKGGGVG 221
Score = 31.2 bits (69), Expect = 0.17
Identities = 23/59 (38%), Positives = 27/59 (44%), Gaps = 6/59 (10%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRG-GYPGNRGGNYGGGY--PGNRGGNYCRYGCCG 105
K G GGG G GG GGG + G G +GG GGG G GG + + G G
Sbjct: 186 KGGGIGGGI---GKGGGIGGGIGKGGGVGGGFGKGGGVGGGIGKGGGVGGGFGKGGGVG 241
Score = 31.2 bits (69), Expect = 0.17
Identities = 21/48 (43%), Positives = 24/48 (49%), Gaps = 4/48 (8%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
+G GG G G GG GGG + GG G +GG GGG G GG
Sbjct: 234 FGKGGGVGGGIGKGGGIGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 279
Score = 30.8 bits (68), Expect = 0.22
Identities = 21/48 (43%), Positives = 24/48 (49%), Gaps = 4/48 (8%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
+G GG G G GG GGG + GG G +GG GGG G GG
Sbjct: 124 FGKGGGIGGGIGKGGGVGGGIGKG-GGIGGGIGKGGGVGGGI-GKGGG 169
Score = 30.0 bits (66), Expect = 0.37
Identities = 21/47 (44%), Positives = 23/47 (48%), Gaps = 4/47 (8%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
G GG G G GG GGG + GG G +GG GGG G GG
Sbjct: 135 GKGGGVGGGIGKGGGIGGGIGKG-GGVGGGIGKGGGIGGGI-GKGGG 179
Score = 30.0 bits (66), Expect = 0.37
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
K G GGG G GG GGG + GG G +GG GGG G GG
Sbjct: 266 KGGGIGGGI---GKGGGIGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 309
Score = 30.0 bits (66), Expect = 0.37
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
K G GGG G GG GGG + GG G +GG GGG G GG
Sbjct: 256 KGGGIGGGI---GKGGGIGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 299
Score = 30.0 bits (66), Expect = 0.37
Identities = 21/47 (44%), Positives = 23/47 (48%), Gaps = 4/47 (8%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
G GG G G GG GGG + GG G +GG GGG G GG
Sbjct: 155 GKGGGVGGGIGKGGGIGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 199
Score = 30.0 bits (66), Expect = 0.37
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
K G GGG G GG GGG + GG G +GG GGG G GG
Sbjct: 146 KGGGIGGGI---GKGGGVGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 189
Score = 30.0 bits (66), Expect = 0.37
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
K G GGG G GG GGG + GG G +GG GGG G GG
Sbjct: 246 KGGGIGGGI---GKGGGIGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 289
Score = 30.0 bits (66), Expect = 0.37
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Query: 50 KYGSYGGGYPGRGGGGNYGGGYPRNRGGYPG--NRGGNYGGGYPGNRGG 96
K G GGG G GG GGG + GG G +GG GGG G GG
Sbjct: 276 KGGGIGGGI---GKGGGIGGGIGKG-GGIGGGIGKGGGIGGGI-GKGGG 319
Score = 29.6 bits (65), Expect = 0.48
Identities = 19/42 (45%), Positives = 20/42 (47%), Gaps = 2/42 (4%)
Query: 55 GGGYPGR-GGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG 95
GGG G G GG GGG + GG G G GGG G G
Sbjct: 297 GGGIGGGIGKGGGIGGGIGKG-GGIGGGGGFGKGGGIGGGIG 337
Score = 27.3 bits (59), Expect = 2.4
Identities = 22/57 (38%), Positives = 25/57 (43%), Gaps = 10/57 (17%)
Query: 50 KYGSYGGGYP--GR-----GGGGNYGGGYPRNRGGYPG---NRGGNYGGGYPGNRGG 96
K G GGG G G GG GGG +GG G +GG GGG +GG
Sbjct: 296 KGGGIGGGIGKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGGGFGKGG 352
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 49.3 bits (116), Expect = 6e-07
Identities = 38/101 (37%), Positives = 44/101 (42%), Gaps = 17/101 (16%)
Query: 22 EVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRGG---------GGNYGGGYP 72
E + RD E + KE D + +N+A+ GGG GRGG GG Y GG
Sbjct: 57 EKAMRDAIEEMNG--KELDGRVITVNEAQSRGSGGGGGGRGGSGGGYRSGGGGGYSGG-- 112
Query: 73 RNRGGYPGNRGGNY---GGGYPGNRGGNYCRYGCCGDRYYG 110
GGY G GG Y GGY GG YG G R G
Sbjct: 113 -GGGGYSGGGGGGYERRSGGYGSGGGGGGRGYGGGGRREGG 152
Score = 49.3 bits (116), Expect = 6e-07
Identities = 24/45 (53%), Positives = 27/45 (59%), Gaps = 2/45 (4%)
Query: 48 DAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPG 92
+ + G YG G G GGG YGGG R GGY G GG+YGGG G
Sbjct: 126 ERRSGGYGSG--GGGGGRGYGGGGRREGGGYGGGDGGSYGGGGGG 168
Score = 44.7 bits (104), Expect = 1e-05
Identities = 26/59 (44%), Positives = 27/59 (45%), Gaps = 17/59 (28%)
Query: 55 GGGYPGRGGGG---NYGGGYPRNRGGYPGN--------------RGGNYGGGYPGNRGG 96
GGGY G GGGG GGGY R GGY GG YGGG G+ GG
Sbjct: 106 GGGYSGGGGGGYSGGGGGGYERRSGGYGSGGGGGGRGYGGGGRREGGGYGGGDGGSYGG 164
Score = 44.3 bits (103), Expect = 2e-05
Identities = 26/57 (45%), Positives = 27/57 (46%), Gaps = 12/57 (21%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGG----------NYGGGYPGNRGGNY 98
G GGG G GGG GGGY R GGY GG GGGY G GG+Y
Sbjct: 108 GYSGGGGGGYSGGG--GGGYERRSGGYGSGGGGGGRGYGGGGRREGGGYGGGDGGSY 162
>At4g38680 glycine-rich protein 2 (GRP2)
Length = 203
Score = 49.3 bits (116), Expect = 6e-07
Identities = 34/92 (36%), Positives = 36/92 (38%), Gaps = 27/92 (29%)
Query: 51 YGSYGGGYPGRGGGGNYG--------------------GGYPRNRGGYPGNRG-----GN 85
YG GGGY GRGGGG G GGY GGY G G G
Sbjct: 111 YGGGGGGYGGRGGGGRGGSDCYKCGEPGHMARDCSEGGGGYGGGGGGYGGGGGYGGGGGG 170
Query: 86 YGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCS 117
YGGG G GG Y C ++ R C S
Sbjct: 171 YGGGGRGGGGGGGSCYSCGESGHF--ARDCTS 200
Score = 48.9 bits (115), Expect = 8e-07
Identities = 38/104 (36%), Positives = 50/104 (47%), Gaps = 8/104 (7%)
Query: 19 ISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG-RGG-GGNYGGGYPRNRG 76
+++E + E +N + +A D + G+ GGG G RGG GG GGG R G
Sbjct: 51 LAAEEAVEFEVEIDNNNRPKAIDVSGPDGAPVQGNSGGGSSGGRGGFGGGRGGG--RGSG 108
Query: 77 GYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAG 120
G G GG YGG G RGG+ C CG+ G + CS G
Sbjct: 109 GGYGGGGGGYGGRGGGGRGGSDCYK--CGEP--GHMARDCSEGG 148
Score = 40.8 bits (94), Expect = 2e-04
Identities = 28/58 (48%), Positives = 30/58 (51%), Gaps = 7/58 (12%)
Query: 55 GGGYPGRGGG--GNYGGGYPRNRGGY-PGNRGGNYGGG--YPGNRGGNYCRYGCCGDR 107
GGGY G GGG G GGGY GGY G RGG GGG Y G++ R G R
Sbjct: 148 GGGYGGGGGGYGG--GGGYGGGGGGYGGGGRGGGGGGGSCYSCGESGHFARDCTSGGR 203
>At5g46730 unknown protein
Length = 268
Score = 48.9 bits (115), Expect = 8e-07
Identities = 26/49 (53%), Positives = 27/49 (55%), Gaps = 3/49 (6%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRG-GYPGNRGGNYGGGYPGNRGGNY 98
YG GGG G GGGG YGGG G G G GG YGGG G GG +
Sbjct: 209 YGGGGGG--GSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGAAGGYGGGH 255
Score = 46.2 bits (108), Expect = 5e-06
Identities = 28/66 (42%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Query: 51 YGSYGG-GYPGRGGGGNYGGGYPRNRG-----GYPGNRGGNYGGGYPGNRGGNYCRYGCC 104
Y S GG G+ G GGG GGY G GY G GG+ GGG G+ GG YG
Sbjct: 79 YASGGGSGHGGGGGGAASSGGYASGAGEGGGGGYGGAAGGHAGGGGGGSGGGGGSAYGAG 138
Query: 105 GDRYYG 110
G+ G
Sbjct: 139 GEHASG 144
Score = 45.4 bits (106), Expect = 8e-06
Identities = 22/40 (55%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query: 52 GSYGGGYPGRGG-GGNYGGGYPRNRGGYPGNRGGNYGGGY 90
G++GGGY GG GG YGGG GG G GG+ GGGY
Sbjct: 227 GAHGGGYGSGGGEGGGYGGGAAGGYGGGHGGGGGHGGGGY 266
Score = 44.7 bits (104), Expect = 1e-05
Identities = 29/64 (45%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Query: 52 GSYGG-GYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYP----GNRGGNYCRYGCCGD 106
G++GG GY G GGG GGG GGY G GG GGG G GG Y G G
Sbjct: 182 GAHGGSGYGGGEGGGAGGGGSHGGAGGYGGGGGGGSGGGGAYGGGGAHGGGYGSGGGEGG 241
Query: 107 RYYG 110
Y G
Sbjct: 242 GYGG 245
Score = 44.7 bits (104), Expect = 1e-05
Identities = 25/52 (48%), Positives = 26/52 (49%), Gaps = 4/52 (7%)
Query: 47 NDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
N YG GG G GGGG GG + GGY G GG YGGG GG Y
Sbjct: 26 NLLNYGQASGG-GGHGGGGGSGG---VSSGGYGGESGGGYGGGSGEGAGGGY 73
Score = 44.3 bits (103), Expect = 2e-05
Identities = 24/47 (51%), Positives = 26/47 (55%), Gaps = 5/47 (10%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPR---NRGGYPGNRGGNYGGGYPGNRG 95
GS GGG GGGG +GGGY GGY G G YGGG+ G G
Sbjct: 216 GSGGGG--AYGGGGAHGGGYGSGGGEGGGYGGGAAGGYGGGHGGGGG 260
Score = 42.4 bits (98), Expect = 7e-05
Identities = 26/52 (50%), Positives = 27/52 (51%), Gaps = 6/52 (11%)
Query: 52 GSYGG--GYPGRGGGGNYGGGYPRNRGGYPG---NRGGNYGGGYPGNRGGNY 98
GS+GG GY G GGGG GGG GG G GG GGGY G G Y
Sbjct: 201 GSHGGAGGYGG-GGGGGSGGGGAYGGGGAHGGGYGSGGGEGGGYGGGAAGGY 251
Score = 41.6 bits (96), Expect = 1e-04
Identities = 23/49 (46%), Positives = 26/49 (52%), Gaps = 3/49 (6%)
Query: 52 GSYGG--GYPGRGGGGNYGGGY-PRNRGGYPGNRGGNYGGGYPGNRGGN 97
G YGG GY GG G+ GGG + GGY G GGGY G GG+
Sbjct: 71 GGYGGAEGYASGGGSGHGGGGGGAASSGGYASGAGEGGGGGYGGAAGGH 119
Score = 41.2 bits (95), Expect = 2e-04
Identities = 23/44 (52%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query: 55 GGGYPGRG-GGGNYGGGYPRNRGGYPGNRGGNYGG-GYPGNRGG 96
GGG G GGG YGGG GG G+ GG +GG GY G GG
Sbjct: 152 GGGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGG 195
Score = 41.2 bits (95), Expect = 2e-04
Identities = 21/45 (46%), Positives = 23/45 (50%), Gaps = 2/45 (4%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G+YGGG GGGG G GY G GG GGG G+ GG
Sbjct: 163 GAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGGG--GSHGG 205
Score = 40.0 bits (92), Expect = 4e-04
Identities = 30/79 (37%), Positives = 33/79 (40%), Gaps = 8/79 (10%)
Query: 52 GSYGGGYPGRGGGGNYGG-GYPRNRGGYPGNRG-----GNYGGGYPGNRGGN--YCRYGC 103
G +GGG G GG +GG GY GG G G G YGGG G GG Y G
Sbjct: 169 GGHGGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSHGGAGGYGGGGGGGSGGGGAYGGGGA 228
Query: 104 CGDRYYGSCRKCCSYAGEA 122
G Y + Y G A
Sbjct: 229 HGGGYGSGGGEGGGYGGGA 247
Score = 38.1 bits (87), Expect = 0.001
Identities = 19/46 (41%), Positives = 21/46 (45%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGN 97
GS GGG G GG + GY G G YGGG G GG+
Sbjct: 126 GSGGGGGSAYGAGGEHASGYGNGAGEGGGAGASGYGGGAYGGGGGH 171
Score = 38.1 bits (87), Expect = 0.001
Identities = 21/45 (46%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGY G GGG YGGG GG G G GG G+ GG
Sbjct: 47 GVSSGGYGGESGGG-YGGGSGEGAGGGYGGAEGYASGGGSGHGGG 90
Score = 36.6 bits (83), Expect = 0.004
Identities = 24/65 (36%), Positives = 26/65 (39%), Gaps = 11/65 (16%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGG----------NYGGGYPGNRGGNYCR 100
YG+ G G G G YGGG GG+ G GG YGGG G GG
Sbjct: 145 YGN-GAGEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGGGGSH 203
Query: 101 YGCCG 105
G G
Sbjct: 204 GGAGG 208
Score = 36.6 bits (83), Expect = 0.004
Identities = 21/48 (43%), Positives = 22/48 (45%), Gaps = 5/48 (10%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGN----YGGGYPGNRG 95
G YGGG G G GG YGG GG G+ GG GGY G
Sbjct: 59 GGYGGG-SGEGAGGGYGGAEGYASGGGSGHGGGGGGAASSGGYASGAG 105
Score = 36.2 bits (82), Expect = 0.005
Identities = 22/55 (40%), Positives = 25/55 (45%), Gaps = 7/55 (12%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
YG GG+ G GGGG+ GGG G GG + GY GN G G G
Sbjct: 112 YGGAAGGHAGGGGGGSGGGG------GSAYGAGGEHASGY-GNGAGEGGGAGASG 159
Score = 33.5 bits (75), Expect = 0.033
Identities = 20/48 (41%), Positives = 24/48 (49%), Gaps = 2/48 (4%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRN--RGGYPGNRGGNYGGGYPGNRGG 96
YG+ G G G G GGG + GG G GG+ GGG G+ GG
Sbjct: 135 YGAGGEHASGYGNGAGEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGG 182
Score = 32.0 bits (71), Expect = 0.097
Identities = 17/44 (38%), Positives = 19/44 (42%)
Query: 55 GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
GGG GGGG+ G + GY G G G G GG Y
Sbjct: 122 GGGGGSGGGGGSAYGAGGEHASGYGNGAGEGGGAGASGYGGGAY 165
>At2g36120 unknown protein
Length = 255
Score = 48.5 bits (114), Expect = 1e-06
Identities = 30/67 (44%), Positives = 34/67 (49%), Gaps = 10/67 (14%)
Query: 52 GSYGGGY---PGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGN-----RGGNYCRYGC 103
G++GGGY G G GG YGGG GG G GGN GGG G+ GG Y G
Sbjct: 141 GAHGGGYGGGQGAGAGGGYGGGGAGGHGG--GGGGGNGGGGGGGSGEGGAHGGGYGAGGG 198
Query: 104 CGDRYYG 110
G+ Y G
Sbjct: 199 AGEGYGG 205
Score = 47.8 bits (112), Expect = 2e-06
Identities = 24/47 (51%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G YGGG G GGG YGGG GG G G GGG G GG Y
Sbjct: 102 GGYGGG-SGEGGGAGYGGGEAGGHGGGGGGGAGGGGGGGGGAHGGGY 147
Score = 45.4 bits (106), Expect = 8e-06
Identities = 23/45 (51%), Positives = 24/45 (53%), Gaps = 3/45 (6%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G G G G GGGG +GGGY GG GG YGGG G GG
Sbjct: 128 GGGGAGGGGGGGGGAHGGGY---GGGQGAGAGGGYGGGGAGGHGG 169
Score = 45.1 bits (105), Expect = 1e-05
Identities = 22/47 (46%), Positives = 26/47 (54%), Gaps = 1/47 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G +GGG G G GG GGG + GGY G +G GGGY G G +
Sbjct: 122 GGHGGG-GGGGAGGGGGGGGGAHGGGYGGGQGAGAGGGYGGGGAGGH 167
Score = 45.1 bits (105), Expect = 1e-05
Identities = 25/54 (46%), Positives = 26/54 (47%), Gaps = 5/54 (9%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
G GG+ G GGGG GGG GG G GG YGGG GG Y G G
Sbjct: 118 GGEAGGHGGGGGGGAGGGG-----GGGGGAHGGGYGGGQGAGAGGGYGGGGAGG 166
Score = 44.7 bits (104), Expect = 1e-05
Identities = 24/49 (48%), Positives = 26/49 (52%), Gaps = 4/49 (8%)
Query: 52 GSYGGGYPGRGGGG---NYGGGYPRNRG-GYPGNRGGNYGGGYPGNRGG 96
G +GGG G GGGG YGGG G GY G G +GGG G GG
Sbjct: 86 GGHGGGAGGGGGGGPGGGYGGGSGEGGGAGYGGGEAGGHGGGGGGGAGG 134
Score = 43.5 bits (101), Expect = 3e-05
Identities = 22/47 (46%), Positives = 25/47 (52%), Gaps = 3/47 (6%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G +G G+ G GGGG +GGG GG PG G YGGG G Y
Sbjct: 73 GGHGEGHIGGGGGGGHGGGAGGGGGGGPG---GGYGGGSGEGGGAGY 116
Score = 43.1 bits (100), Expect = 4e-05
Identities = 22/47 (46%), Positives = 25/47 (52%), Gaps = 1/47 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G++GGGY G GGG G G GG+ G GG G G G GG Y
Sbjct: 187 GAHGGGY-GAGGGAGEGYGGGAGAGGHGGGGGGGGGSGGGGGGGGGY 232
Score = 42.0 bits (97), Expect = 9e-05
Identities = 21/45 (46%), Positives = 24/45 (52%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G+ GGG G G GG +GGGY G G GG GG+ G GG
Sbjct: 174 GNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGG 218
Score = 40.4 bits (93), Expect = 3e-04
Identities = 23/46 (50%), Positives = 24/46 (52%), Gaps = 2/46 (4%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNR-GGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGG YGGG GGY G G +GGG G GG
Sbjct: 133 GGGGGGGGGAHGGG-YGGGQGAGAGGGYGGGGAGGHGGGGGGGNGG 177
Score = 40.4 bits (93), Expect = 3e-04
Identities = 21/47 (44%), Positives = 22/47 (46%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G GGG G GGG GGG GGY G G G GY G G +
Sbjct: 78 GHIGGGGGGGHGGGAGGGGGGGPGGGYGGGSGEGGGAGYGGGEAGGH 124
Score = 40.4 bits (93), Expect = 3e-04
Identities = 25/52 (48%), Positives = 26/52 (49%), Gaps = 6/52 (11%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYG 102
YG G G G GGGG GGG GG GG +GGGY G GG YG
Sbjct: 159 YGGGGAGGHGGGGGGGNGGG-----GGGGSGEGGAHGGGY-GAGGGAGEGYG 204
Score = 40.0 bits (92), Expect = 4e-04
Identities = 22/42 (52%), Positives = 24/42 (56%), Gaps = 2/42 (4%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRG-GYPGNRGGNYG-GGY 90
+G GGG G GGGG GGGY G G+ G GG G GGY
Sbjct: 212 HGGGGGGGGGSGGGGGGGGGYAAASGYGHGGGAGGGEGSGGY 253
Score = 39.7 bits (91), Expect = 5e-04
Identities = 21/48 (43%), Positives = 24/48 (49%), Gaps = 1/48 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG-GNY 98
G+ GG+ G GGGG GG GGY G +GGG G G G Y
Sbjct: 206 GAGAGGHGGGGGGGGGSGGGGGGGGGYAAASGYGHGGGAGGGEGSGGY 253
Score = 38.5 bits (88), Expect = 0.001
Identities = 23/46 (50%), Positives = 25/46 (54%), Gaps = 4/46 (8%)
Query: 54 YGGGYPGRGGG-GNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
YGGG G GGG G +G G+ GG G GG GGG G GG Y
Sbjct: 62 YGGG-SGEGGGAGGHGEGHIGGGGG--GGHGGGAGGGGGGGPGGGY 104
Score = 37.7 bits (86), Expect = 0.002
Identities = 22/64 (34%), Positives = 30/64 (46%)
Query: 33 SNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPG 92
S+++ EA+ +N G G G GGG+ GG GG G+ G+ GGG G
Sbjct: 27 SSSESEAEVAAYGVNSGLSAGLGVGIGGGPGGGSGYGGGSGEGGGAGGHGEGHIGGGGGG 86
Query: 93 NRGG 96
GG
Sbjct: 87 GHGG 90
Score = 37.0 bits (84), Expect = 0.003
Identities = 25/66 (37%), Positives = 27/66 (40%), Gaps = 7/66 (10%)
Query: 52 GSYGGGYPGRGG--GGNYGGGYPRNRG-----GYPGNRGGNYGGGYPGNRGGNYCRYGCC 104
G GGG G GG GG YG G G G G+ GG GGG G GG Y
Sbjct: 176 GGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGSGGGGGGGGGYAAA 235
Query: 105 GDRYYG 110
+G
Sbjct: 236 SGYGHG 241
Score = 36.6 bits (83), Expect = 0.004
Identities = 19/45 (42%), Positives = 20/45 (44%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG GG G GG G G GG +GGG G GG
Sbjct: 54 GGPGGGSGYGGGSGEGGGAGGHGEGHIGGGGGGGHGGGAGGGGGG 98
Score = 35.8 bits (81), Expect = 0.007
Identities = 19/45 (42%), Positives = 20/45 (44%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG 95
YG+ GG G GGG GG GG GG GGGY G
Sbjct: 193 YGAGGGAGEGYGGGAGAGGHGGGGGGGGGSGGGGGGGGGYAAASG 237
Score = 35.4 bits (80), Expect = 0.009
Identities = 24/56 (42%), Positives = 25/56 (43%), Gaps = 4/56 (7%)
Query: 51 YGSYGGGYPGRGGGGN---YGGGYPRNRGGYPGNRGGNYGGGYPGNRG-GNYCRYG 102
YG G G GG G GGG + GG G GG GGGY G G G YG
Sbjct: 62 YGGGSGEGGGAGGHGEGHIGGGGGGGHGGGAGGGGGGGPGGGYGGGSGEGGGAGYG 117
Score = 34.3 bits (77), Expect = 0.019
Identities = 22/52 (42%), Positives = 22/52 (42%), Gaps = 8/52 (15%)
Query: 52 GSYGGGYPGR-----GGGGNYGGGYPRNRGGYPG---NRGGNYGGGYPGNRG 95
G YGGG G GGG GGG GG G GG G GY G G
Sbjct: 157 GGYGGGGAGGHGGGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAG 208
Score = 32.0 bits (71), Expect = 0.097
Identities = 19/44 (43%), Positives = 20/44 (45%), Gaps = 1/44 (2%)
Query: 54 YGGGYPGRG-GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
YGGG G GGG GGG GG G G G+ G GG
Sbjct: 203 YGGGAGAGGHGGGGGGGGGSGGGGGGGGGYAAASGYGHGGGAGG 246
>At2g32690 unknown protein
Length = 201
Score = 47.8 bits (112), Expect = 2e-06
Identities = 26/59 (44%), Positives = 28/59 (47%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYG 110
G GGGY G GGG GGG GG+ G GG +GGG G G G G Y G
Sbjct: 139 GGAGGGYGGGAGGGLGGGGGIGGGGGFGGGGGGGFGGGAGGGFGKGIGGGGGLGGGYVG 197
Score = 45.1 bits (105), Expect = 1e-05
Identities = 24/45 (53%), Positives = 24/45 (53%), Gaps = 5/45 (11%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG GG G GG YGGG G GG
Sbjct: 116 GLGGGGGLGGGGGGGLGGG-----GGLGGGAGGGYGGGAGGGLGG 155
Score = 42.4 bits (98), Expect = 7e-05
Identities = 24/45 (53%), Positives = 24/45 (53%), Gaps = 2/45 (4%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG GG G GG GGGY G GG
Sbjct: 109 GGLGGG-SGLGGGGGLGGGGGGGLGG-GGGLGGGAGGGYGGGAGG 151
Score = 42.0 bits (97), Expect = 9e-05
Identities = 24/49 (48%), Positives = 26/49 (52%), Gaps = 2/49 (4%)
Query: 52 GSYGGGYPGRGG-GGNYGGGYPRNRGGYPGNRGG-NYGGGYPGNRGGNY 98
G GGG G GG GG GGGY GG G GG GGG+ G GG +
Sbjct: 125 GGGGGGLGGGGGLGGGAGGGYGGGAGGGLGGGGGIGGGGGFGGGGGGGF 173
Score = 41.6 bits (96), Expect = 1e-04
Identities = 23/47 (48%), Positives = 24/47 (50%), Gaps = 3/47 (6%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
G GGG G GGGG GGG GGY G GG GGG GG +
Sbjct: 122 GLGGGGGGGLGGGGGLGGGAG---GGYGGGAGGGLGGGGGIGGGGGF 165
Score = 40.8 bits (94), Expect = 2e-04
Identities = 23/46 (50%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPG-NRGGNYGGGYPGNRGG 96
G GGG G G GG YGGG GG G GG +GGG G GG
Sbjct: 130 GLGGGGGLGGGAGGGYGGGAGGGLGGGGGIGGGGGFGGGGGGGFGG 175
Score = 39.7 bits (91), Expect = 5e-04
Identities = 24/50 (48%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGG---NYGGGYPGNRGGNY 98
G GGG G GGGG GGG GG G GG GGG G GG Y
Sbjct: 97 GGLGGG-SGLGGGGGLGGGSGLGGGGGLGGGGGGGLGGGGGLGGGAGGGY 145
Score = 38.1 bits (87), Expect = 0.001
Identities = 22/47 (46%), Positives = 27/47 (56%), Gaps = 10/47 (21%)
Query: 55 GGGYPGRGGGG---NYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
GGG+ G GGGG GGG+ + GG GG GGGY G GG++
Sbjct: 162 GGGFGGGGGGGFGGGAGGGFGKGIGG-----GGGLGGGYVG--GGHH 201
Score = 37.4 bits (85), Expect = 0.002
Identities = 27/64 (42%), Positives = 27/64 (42%), Gaps = 6/64 (9%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPG-----NRGGNYGGGYPGNRGGNYCRYGCCGD 106
G GGG G GGGG GGG GG G GG GGG G GG G G
Sbjct: 85 GGLGGG-GGLGGGGGLGGGSGLGGGGGLGGGSGLGGGGGLGGGGGGGLGGGGGLGGGAGG 143
Query: 107 RYYG 110
Y G
Sbjct: 144 GYGG 147
Score = 37.0 bits (84), Expect = 0.003
Identities = 20/46 (43%), Positives = 23/46 (49%), Gaps = 1/46 (2%)
Query: 51 YGSYGGGYPGRGGGGNYGGGY-PRNRGGYPGNRGGNYGGGYPGNRG 95
YG GG G GGG GGG+ GG+ G GG +G G G G
Sbjct: 145 YGGGAGGGLGGGGGIGGGGGFGGGGGGGFGGGAGGGFGKGIGGGGG 190
Score = 36.6 bits (83), Expect = 0.004
Identities = 21/46 (45%), Positives = 22/46 (47%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
+G GG G GGGG GGG GG G GG GGG G G
Sbjct: 59 FGKGLGGGGGLGGGGGLGGGGGLGGGGGLGGGGGLGGGGGLGGGSG 104
Score = 36.6 bits (83), Expect = 0.004
Identities = 22/45 (48%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG GG G GG GG G GG
Sbjct: 67 GGLGGG-GGLGGGGGLGGGGGLGGGGGLGGGGGLGGGSGLGGGGG 110
Score = 34.7 bits (78), Expect = 0.015
Identities = 23/48 (47%), Positives = 25/48 (51%), Gaps = 4/48 (8%)
Query: 50 KYGSYGGGY-PGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
K+G Y G+ G GGGG GGG GG G GG GGG G GG
Sbjct: 51 KHGIYKKGFGKGLGGGGGLGGGGGLGGGGGLGG-GGGLGGG--GGLGG 95
Score = 34.3 bits (77), Expect = 0.019
Identities = 21/45 (46%), Positives = 21/45 (46%), Gaps = 1/45 (2%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
G GGG G GGGG GGG GG G G GGG G G
Sbjct: 73 GGLGGG-GGLGGGGGLGGGGGLGGGGGLGGGSGLGGGGGLGGGSG 116
Score = 34.3 bits (77), Expect = 0.019
Identities = 21/44 (47%), Positives = 21/44 (47%), Gaps = 6/44 (13%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRG 95
G GGG G GGGG GGG GG G G GGG G G
Sbjct: 79 GGLGGG-GGLGGGGGLGGG-----GGLGGGSGLGGGGGLGGGSG 116
Score = 28.5 bits (62), Expect = 1.1
Identities = 20/44 (45%), Positives = 22/44 (49%), Gaps = 1/44 (2%)
Query: 55 GGGYPGRGG-GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGN 97
GGG G GG GG G G GG G GG+ GG G GG+
Sbjct: 72 GGGLGGGGGLGGGGGLGGGGGLGGGGGLGGGSGLGGGGGLGGGS 115
Score = 28.5 bits (62), Expect = 1.1
Identities = 20/43 (46%), Positives = 20/43 (46%), Gaps = 1/43 (2%)
Query: 55 GGGYPGRGG-GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGG 96
GGG G GG GG G G GG G GG GG G GG
Sbjct: 78 GGGLGGGGGLGGGGGLGGGGGLGGGSGLGGGGGLGGGSGLGGG 120
>At1g75550 hypothetical protein
Length = 167
Score = 47.8 bits (112), Expect = 2e-06
Identities = 37/116 (31%), Positives = 51/116 (43%), Gaps = 16/116 (13%)
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYG-------- 52
M + +++++ LL +I + VS D SN Q + N+ N G
Sbjct: 1 MKTISSLVVIFLLILIFQTNKLVSGHD----RSNDQDTVKNNNNDENSTTTGVVVRDKKR 56
Query: 53 SYGGG---YPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCG 105
SY GG Y GGG GGG GG G GG +G G G GG + ++GC G
Sbjct: 57 SYNGGSGSYRWGWGGGGGGGGGGGGGGGGGGGGGGGWGWG-GGGGGGGWYKWGCGG 111
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,833,847
Number of Sequences: 26719
Number of extensions: 229042
Number of successful extensions: 6901
Number of sequences better than 10.0: 361
Number of HSP's better than 10.0 without gapping: 246
Number of HSP's successfully gapped in prelim test: 116
Number of HSP's that attempted gapping in prelim test: 805
Number of HSP's gapped (non-prelim): 2588
length of query: 133
length of database: 11,318,596
effective HSP length: 88
effective length of query: 45
effective length of database: 8,967,324
effective search space: 403529580
effective search space used: 403529580
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 54 (25.4 bits)
Medicago: description of AC138451.12


