

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.6 + phase: 0
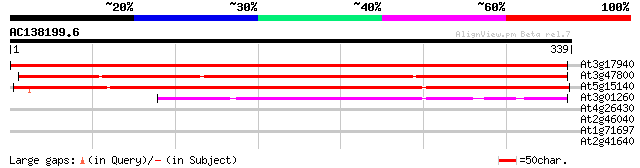
(339 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g17940 aldose 1-epimerase like protein 530 e-151
At3g47800 aldose 1-epimerase - like protein 332 2e-91
At5g15140 putative aldose 1-epimerase - like protein 311 4e-85
At3g01260 putative aldose 1-epimerase 157 8e-39
At4g26430 COP9 signalosome subunit 6 (CSN6B) 30 2.5
At2g46040 unknown protein 29 4.2
At1g71697 choline kinase-like protein 28 7.2
At2g41640 unknown protein 28 9.3
>At3g17940 aldose 1-epimerase like protein
Length = 341
Score = 530 bits (1364), Expect = e-151
Identities = 254/340 (74%), Positives = 293/340 (85%), Gaps = 3/340 (0%)
Query: 1 MADQNKK-PEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLA 59
MADQ+K PEIFELNNGTMQ+ ++N G TITS SVP K+G L DVVLG DSV+ Y KGLA
Sbjct: 1 MADQSKNTPEIFELNNGTMQVKISNYGTTITSLSVPDKNGKLGDVVLGFDSVDPYVKGLA 60
Query: 60 PYFGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKK-GET 118
PYFGCIVGRVANRIK GKF+L+GV Y+LP+NK PN+LHGGN GFDKK+W+V +K+ GE
Sbjct: 61 PYFGCIVGRVANRIKEGKFSLNGVNYTLPINKPPNSLHGGNKGFDKKIWEVAGHKRDGEK 120
Query: 119 PSITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLA 178
P ITFKY S DGEEGYPG ++VTATYTLTS+TTMRLDME V +NK T INLAQHTYWNLA
Sbjct: 121 PFITFKYHSADGEEGYPGAVSVTATYTLTSATTMRLDMEAVPENKDTPINLAQHTYWNLA 180
Query: 179 GHSSGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVGLGYD 238
GH SGNILDH I+I +H+TPVD+ TVPTGEI+PVKGTPFDFT EKRIG++I +VG+GYD
Sbjct: 181 GHDSGNILDHKIQIWGSHITPVDEYTVPTGEILPVKGTPFDFTEEKRIGESIGEVGIGYD 240
Query: 239 HNYVLDC-GEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGK 297
HNYVLDC +EK GL+HAAK+ D +SSRVLNLWTN PG+QFYTGNYV+ V GKG AVYGK
Sbjct: 241 HNYVLDCPDQEKEGLKHAAKLSDAASSRVLNLWTNVPGMQFYTGNYVNGVVGKGNAVYGK 300
Query: 298 HAGLCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFS 337
HAG+CLETQGFP+A+N+ NFPSVVVK GEKY H+MLFEFS
Sbjct: 301 HAGVCLETQGFPNAINQSNFPSVVVKAGEKYNHTMLFEFS 340
>At3g47800 aldose 1-epimerase - like protein
Length = 358
Score = 332 bits (850), Expect = 2e-91
Identities = 169/332 (50%), Positives = 211/332 (62%), Gaps = 4/332 (1%)
Query: 6 KKPEIFELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAPYFGCI 65
+K + ++L G++ + TN G +TS +P + G DVVLG D+V+ Y K YFG I
Sbjct: 29 EKIKTYKLTRGSLSVTFTNYGAVMTSLLLPDRHGKQDDVVLGFDTVDGY-KNDTTYFGAI 87
Query: 66 VGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPSITFKY 125
VGRVANRI KF L+G Y N+ NTLHGG+ GF +W V +Y T ITF Y
Sbjct: 88 VGRVANRIGGAKFKLNGHLYKTDPNEGRNTLHGGSKGFSDVIWSVQKYVP--TSHITFTY 145
Query: 126 DSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGHSSGNI 185
DS DGEEG+PG++TV TY L + + ME NKPT INLA HTYWNL H+SGNI
Sbjct: 146 DSFDGEEGFPGNVTVKVTYMLIGENKLGVKMEAKPLNKPTPINLALHTYWNLHSHNSGNI 205
Query: 186 LDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVGLGYDHNYVLDC 245
L H I++ A +TPVD +PTGEI + GTP+DF + IG I+++ GYD NYV+D
Sbjct: 206 LSHKIQLLAGKITPVDDKLIPTGEITSITGTPYDFLEPREIGSRIHELPGGYDINYVID- 264
Query: 246 GEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAGLCLET 305
G LR A V + + R + LWTN PGVQFYT N + V GKG AVY K+ GLCLET
Sbjct: 265 GPIGKHLRKTAVVTEQVTGRKMELWTNQPGVQFYTSNMMKRVVGKGKAVYEKYGGLCLET 324
Query: 306 QGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFS 337
QGFPD+VN NFPS +V PGE Y H MLF F+
Sbjct: 325 QGFPDSVNHKNFPSQIVNPGESYLHVMLFRFT 356
>At5g15140 putative aldose 1-epimerase - like protein
Length = 490
Score = 311 bits (796), Expect = 4e-85
Identities = 160/339 (47%), Positives = 216/339 (63%), Gaps = 6/339 (1%)
Query: 3 DQNKKPEI--FELNNGTMQLLVTNLGCTITSFSVPAKDGVLSDVVLGLDSVESYQKGLAP 60
D N+K +I +EL G + + TN G +I S P K+G + D+VLG DSV++Y+
Sbjct: 153 DDNEKEKIGLYELKKGNLTVKFTNWGASIISLHFPDKNGKMDDIVLGYDSVKTYKTDKV- 211
Query: 61 YFGCIVGRVANRIKNGKFTLDGVEYSLPLNKAPNTLHGGNVGFDKKVWDVLEYK-KGETP 119
YFG VGRVANRI GKF L+G EY +N NTLHGG GF VW V +++ G+ P
Sbjct: 212 YFGATVGRVANRIGKGKFKLNGKEYKTSVNDGKNTLHGGKKGFGDVVWAVAKHQYDGKKP 271
Query: 120 SITFKYDSHDGEEGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPTIINLAQHTYWNLAG 179
I F + S DG++G+PG+++VT TY L + + ME K+K T +NLA H+YWNL G
Sbjct: 272 HIVFTHTSPDGDQGFPGELSVTVTYKLVKDNELSVVMEAKPKDKATPVNLAHHSYWNLGG 331
Query: 180 HSSGNILDHSIKISANHVTPVDQNTVPTGEIVPVKGTPFDFTSEKRIGDTINQVGLGYDH 239
H+SG+IL I+I + TPVD +PTG+I PVKGT +DF + I D + + GYD
Sbjct: 332 HNSGDILSEEIQILGSGYTPVDGELIPTGKINPVKGTAYDFLQLRPIKDNMKDLKTGYDI 391
Query: 240 NYVLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHA 299
NY LD +K +R ++ D S R + L N G+QFYTG + +V GK GAVY
Sbjct: 392 NYCLDGKAKK--MRKIVELVDKKSGRKMELSGNQAGLQFYTGGMLKDVKGKNGAVYQAFG 449
Query: 300 GLCLETQGFPDAVNKPNFPSVVVKPGEKYQHSMLFEFSI 338
GLCLETQ +PDA+N P FPS +V+PG+KY+H+MLF+FSI
Sbjct: 450 GLCLETQSYPDALNHPKFPSQIVEPGKKYKHTMLFKFSI 488
>At3g01260 putative aldose 1-epimerase
Length = 378
Score = 157 bits (397), Expect = 8e-39
Identities = 93/250 (37%), Positives = 137/250 (54%), Gaps = 17/250 (6%)
Query: 90 NKAPNTLHGGNVGFDKKVWDVLEYK-KGETPSITFKYD-SHDGEEGYPGDITVTATYTLT 147
N NT+HGG +W V ++K G+ P I F Y S DG++G + VT TY L
Sbjct: 141 NDGKNTIHGGTKESSDVIWRVKKHKHNGKKPYIVFTYTTSPDGDQG---KLEVTVTYKLV 197
Query: 148 SSTTMRLDMEGVAKNKPTIINLAQHTYWNLAGHSSGNILDHSIKISANHVTPVDQNTVPT 207
+++ M+ AK K T +NL +YWNL GH++ +I I+I + T +D N PT
Sbjct: 198 GENRLKMVMQAKAKEKTTPLNLVHRSYWNLGGHNNEDIFSEEIQILGSGYTHLDDNLTPT 257
Query: 208 GEIVPVKGTPFDFTSEKRIGDTINQVGLGYDHNYVLDCGEEKAGLRHAAKVRDPSSSRVL 267
G+I+ VKGTPFDF + I D IN++ GY NY LD K +R ++ D S +
Sbjct: 258 GKILAVKGTPFDFRQLRPIKDNINELKTGYGINYCLDGVANK--MRKVVELVDNKSKIKM 315
Query: 268 NLWTNAPGVQFYTGNYVDNVTGKGGAVYGKHAGLCLETQGFPDAVNKPNFPSVVVKPGEK 327
L T+ G++ T K G+V+ ++GLCLE+ A+N S +++PGE
Sbjct: 316 ELSTDQSGLRLKT------TKPKNGSVHKAYSGLCLESH----AINYQYRSSQIIEPGET 365
Query: 328 YQHSMLFEFS 337
Y+H+MLF+FS
Sbjct: 366 YKHTMLFKFS 375
>At4g26430 COP9 signalosome subunit 6 (CSN6B)
Length = 317
Score = 29.6 bits (65), Expect = 2.5
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 10/81 (12%)
Query: 132 EGYPGDITVTATYTLTSSTTMRLDMEGVAKNKPT-----IINLAQHTYWNLAG-HSSGNI 185
+G P I V +YT+ + R+ ++ VA KP+ LA H L G HS+ +
Sbjct: 166 DGIPQSIFVHTSYTIETVEAERISVDHVAHLKPSDGGSAATQLAAH----LTGIHSAIKM 221
Query: 186 LDHSIKISANHVTPVDQNTVP 206
L+ I++ H+ + + P
Sbjct: 222 LNSRIRVLYQHIVAMQKGDKP 242
>At2g46040 unknown protein
Length = 562
Score = 28.9 bits (63), Expect = 4.2
Identities = 21/90 (23%), Positives = 43/90 (47%), Gaps = 8/90 (8%)
Query: 81 DGVEYS-----LPLNKAPNTLHGGNVGFDKKVWDVLEYKKGETPSITFKYDSHDGEEGYP 135
D V+YS L N++P +++ + GF+KK+ +++ + S ++ S DG P
Sbjct: 11 DAVDYSKTPKSLDANRSPESVNPESTGFEKKIKELISLFRPLLDSFLAEFCSADGFLPLP 70
Query: 136 ---GDITVTATYTLTSSTTMRLDMEGVAKN 162
G+ + L + T + + V++N
Sbjct: 71 AMTGEGRTVDLFNLFLNVTHKGGFDAVSEN 100
>At1g71697 choline kinase-like protein
Length = 346
Score = 28.1 bits (61), Expect = 7.2
Identities = 34/150 (22%), Positives = 58/150 (38%), Gaps = 34/150 (22%)
Query: 69 VANRIKNGKFTLDGVEYSLPLNK------------APNTLHGGNVGFDK--KVWDVLEYK 114
+A+ I+ K+ L+G+E + L + N L GNV D+ +++Y+
Sbjct: 170 LASPIEMDKYRLEGLENEINLLEERLTRDDQEIGFCHNDLQYGNVMIDEVTNAITIIDYE 229
Query: 115 KGETPSITFKYDSHDGE--EGYPGDITVTATYTL-----------------TSSTTMRLD 155
I + +H E Y D YTL T + T +
Sbjct: 230 YSSFNPIAYDIANHFCEMAANYHSDTPHVLDYTLYPGEGERRRFISTYLGSTGNATSDKE 289
Query: 156 MEGVAKNKPTIINLAQHTYWNLAGHSSGNI 185
+E + K+ + LA H +W L G SG++
Sbjct: 290 VERLLKDAESY-TLANHIFWGLWGIISGHV 318
>At2g41640 unknown protein
Length = 500
Score = 27.7 bits (60), Expect = 9.3
Identities = 17/46 (36%), Positives = 24/46 (51%), Gaps = 3/46 (6%)
Query: 242 VLDCGEEKAGLRHAAKVRDPSSSRVLNLWTNAPGVQFYTGNYVDNV 287
V+D +E L K + SS RV +++ + P V F TG Y NV
Sbjct: 153 VMDTVQE---LNLITKDSNKSSDRVCDVYHDVPAVFFSTGGYTGNV 195
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,361,551
Number of Sequences: 26719
Number of extensions: 381822
Number of successful extensions: 563
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 548
Number of HSP's gapped (non-prelim): 8
length of query: 339
length of database: 11,318,596
effective HSP length: 100
effective length of query: 239
effective length of database: 8,646,696
effective search space: 2066560344
effective search space used: 2066560344
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138199.6


