

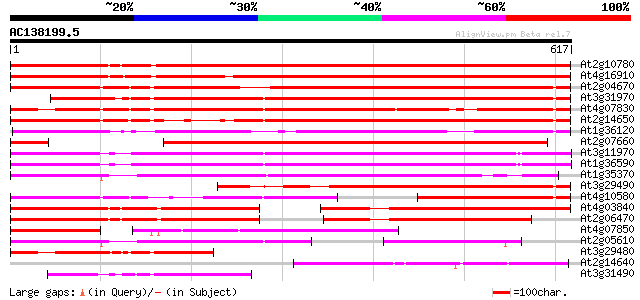
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.5 + phase: 0
(617 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 659 0.0
At4g16910 retrotransposon like protein 632 0.0
At2g04670 putative retroelement pol polyprotein 590 e-169
At3g31970 hypothetical protein 570 e-163
At4g07830 putative reverse transcriptase 541 e-154
At2g14650 putative retroelement pol polyprotein 536 e-152
At1g36120 putative reverse transcriptase gb|AAD22339.1 493 e-139
At2g07660 putative retroelement pol polyprotein 484 e-137
At3g11970 hypothetical protein 377 e-104
At1g36590 hypothetical protein 372 e-103
At1g35370 hypothetical protein 336 2e-92
At3g29490 hypothetical protein 309 3e-84
At4g10580 putative reverse-transcriptase -like protein 274 9e-74
At4g03840 putative transposon protein 270 2e-72
At2g06470 putative retroelement pol polyprotein 269 3e-72
At4g07850 putative polyprotein 212 4e-55
At2g05610 putative retroelement pol polyprotein 202 6e-52
At3g29480 hypothetical protein 178 9e-45
At2g14640 putative retroelement pol polyprotein 175 6e-44
At3g31490 hypothetical protein 168 7e-42
>At2g10780 pseudogene
Length = 1611
Score = 659 bits (1701), Expect = 0.0
Identities = 321/618 (51%), Positives = 441/618 (70%), Gaps = 8/618 (1%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ+G V+AYASRQLR HEKNYPTHDLE+AAVVF LKIWR YLYG++ ++++DHKSLKY+F
Sbjct: 971 MQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIF 1030
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+RQRRW+EL+ DY+ + YHPGKAN VADALSR+ + A + +L+
Sbjct: 1031 TQPELNLRQRRWMELVADYNLDIAYHPGKANQVADALSRRRSEVEAER-SQVDLVNMMGT 1089
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+ V S + LG+ D ++ L I+ AQ+ D + + Q ++++ + G
Sbjct: 1090 LH-VNALSKEVEPLGLGAADQADLLSRIRLAQERDEE----IKGWAQNNKTEYQTSNNGT 1144
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
+ GR+C+P++ +K+ IL E+H+S SIHPG+ KMY DLK+ + W G+K+DVA++V
Sbjct: 1145 IVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAK 1204
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRG-NDAIWVIVDRLTKS 298
C CQ K EHQ P+G++ L +PEWKWD I+MDFVT LP + ++A+WV+VDRLTKS
Sbjct: 1205 CPTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKS 1264
Query: 299 AHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLS 358
AHF+ I+ +AE YI EIV+LHG+P SIVSDRD RFTS+FWK+ Q+ALG+++ LS
Sbjct: 1265 AHFMAISDKDGAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLS 1324
Query: 359 SAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEAL 418
+AYHPQTD QSERTIQ+LED+LR CVL+ GG W+ +L L+EF YNNS+ +SIGM+P+EAL
Sbjct: 1325 TAYHPQTDEQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEAL 1384
Query: 419 YGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQ 478
YGR CR PLCW GER + GP IV +TTE+++ ++ K+K +Q RQK+Y +KRRK+LEFQ
Sbjct: 1385 YGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELEFQ 1444
Query: 479 EGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHV 538
GD V+L+ G GR KKL+P+++GPY+++ERVG VAY++ LPP L+ HNVFHV
Sbjct: 1445 VGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVFHV 1504
Query: 539 SQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGE 598
SQL+K + D ++ +++N+TVE PVRI DR K RGK L++V+W E
Sbjct: 1505 SQLRKCLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMTKGTRGKARDLLKVLWNCRGRE 1564
Query: 599 SLTWELESKMLVSYPELF 616
TWE E+KM ++PE F
Sbjct: 1565 EYTWETENKMKANFPEWF 1582
>At4g16910 retrotransposon like protein
Length = 687
Score = 632 bits (1631), Expect = 0.0
Identities = 312/618 (50%), Positives = 434/618 (69%), Gaps = 16/618 (2%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
+Q+G V+AYASRQLR HEKNYPT+DLE+AAVVF LKIWR YLYG++ ++F+DHKSLKY+F
Sbjct: 63 IQKGSVIAYASRQLRKHEKNYPTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKYIF 122
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+RQRRW++L+ DYD + YHPGKAN V DALSR + A + L+
Sbjct: 123 TQPELNLRQRRWIKLVADYDLNIAYHPGKANQVVDALSRLRTVVEAER-NQVNLVNMMGT 181
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+ L S + LG+ + ++ L I+ AQ+ D + + Q ++++ + G
Sbjct: 182 LHLNA-LSKEVEPLGLRAANQADLLSRIRSAQERDEE----IKGWAQNNKTEYQSSNNGT 236
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
+ GR+C P+++ +K+ IL+E+H+S SIHPG+ KMY DLK+ + W G+K+DVA++V
Sbjct: 237 IVVNGRVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV-- 294
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRG-NDAIWVIVDRLTKS 298
+K EHQ P+GM+ L +PEWKWD I MDFVT LP + ++A+WV+VDRLTKS
Sbjct: 295 ------AKEEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKS 348
Query: 299 AHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLS 358
AHF+ I+ +AE YI EIV+LHG+P SIVSDRD RFTS+FWK Q+ LG+++ LS
Sbjct: 349 AHFMAISDKDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNLS 408
Query: 359 SAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEAL 418
+AYHPQTDGQSERTIQ+LED+LR CVL+ GG W+ +L L+EF YNNS+ +SIGM+P+EAL
Sbjct: 409 TAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEAL 468
Query: 419 YGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQ 478
YGR R PLCW GER + GPA+V +TT+K++ ++ K+K +Q RQK+Y +KRRK+LEFQ
Sbjct: 469 YGRAGRTPLCWTPVGERRLFGPAVVDETTKKMKFLKIKLKEAQDRQKSYANKRRKELEFQ 528
Query: 479 EGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHV 538
GD V+L+ G GR KKL P+++GPY+++ERVG VAY++ LPP L HNVFHV
Sbjct: 529 VGDLVYLKAMTYKGAGRFTSRKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNVFHV 588
Query: 539 SQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGE 598
SQL+K + + ++ +++N+TVE PVRI D+ K RGK + L++++W E
Sbjct: 589 SQLRKCLSEQEESMEDVPPGLKENMTVEAWPVRIMDQMKKGTRGKSMDLLKILWNCGGRE 648
Query: 599 SLTWELESKMLVSYPELF 616
TWE E+KM ++PE F
Sbjct: 649 EYTWETETKMKANFPEWF 666
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 590 bits (1521), Expect = e-169
Identities = 305/619 (49%), Positives = 416/619 (66%), Gaps = 45/619 (7%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ GKV+AYASRQLR HE NYPTHDLE+A V+F LKIWR YLYG + +VF+DHKSLKY+F
Sbjct: 829 MQRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKYIF 888
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
+Q ELN+RQ RW+EL+ DYD + YHPGKANVVADALSRK + A + E L
Sbjct: 889 NQPELNLRQMRWMELVADYDLEIAYHPGKANVVADALSRK--RVGAAPGQSVEALVSEIG 946
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+C + + LG+ +D ++ L + AQ+ D + L+A + E S+++F G
Sbjct: 947 ALRLCVVAREP--LGLEAVDRADLLTRARLAQEKD----EGLIAASKAEGSEYQFAANGT 1000
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
+ GR+C+P +EE+++ IL E+H S SIHPGATKMY DLK+ + W G+KRDVA +V
Sbjct: 1001 IFVYGRVCVPKDEELRREILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWVAE 1060
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSA 299
C VCQ K EHQ P DAIWVI+DRLTKSA
Sbjct: 1061 CDVCQLVKAEHQVP--------------------------------DAIWVIMDRLTKSA 1088
Query: 300 HFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSS 359
HFL I + A LA+ Y+ EIVKLHGVP SIVSDRD +FT FW++ Q +G+K+++S+
Sbjct: 1089 HFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMST 1148
Query: 360 AYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALY 419
AYHPQTDGQSERTIQ+LED+LR+CVL+ GG W HL L+EF YNNSY +SIGMAPFEALY
Sbjct: 1149 AYHPQTDGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALY 1208
Query: 420 GRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQE 479
GR C PL W + ER + G VQ+TTE++++++ +K +Q+RQ++Y DKRR++LEF+
Sbjct: 1209 GRPCWTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEAQARQRSYADKRRRELEFEV 1268
Query: 480 GDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVS 539
GD V+L++ + G R++ KL+P+++GP++I+ERVG VAYR+ LP + H VFHV
Sbjct: 1269 GDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVL 1328
Query: 540 QLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVW--TGATG 597
L+K + V+ ++ N+T+E PVR+ +R++K LR K+IPL++V+W G T
Sbjct: 1329 MLRKCLHKDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLWDCDGVTE 1388
Query: 598 ESLTWELESKMLVSYPELF 616
E TWE E++M + + F
Sbjct: 1389 E--TWEPEARMKARFKKWF 1405
>At3g31970 hypothetical protein
Length = 1329
Score = 570 bits (1469), Expect = e-163
Identities = 290/575 (50%), Positives = 400/575 (69%), Gaps = 20/575 (3%)
Query: 46 RFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMS 105
R +VF+DHKSLKY+F Q ELN+RQRRW+EL+ DYD + YH GKANVVADALSRK + S
Sbjct: 765 RGKVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAYHSGKANVVADALSRKRVGGS 824
Query: 106 A-MMVREFELLEQFRDMSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVA 163
+V E L +C + + LG+ +D ++ L ++ AQ+ D + L+A
Sbjct: 825 VEALVSEIGALR-------LCVMAQEP--LGLEAVDRADLLTRVRLAQEKD----EGLIA 871
Query: 164 RDQTEDSDFKFDDQGVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKI 223
+ + S+++F G + GR+C+P +EE+++ IL E+H S SIHP ATKMY DLK+
Sbjct: 872 ASKADGSEYQFAANGTILVHGRVCVPKDEELRREILSEAHASMFSIHPRATKMYRDLKRY 931
Query: 224 FWWSGLKRDVAQFVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPR 283
+ W G+KRDVA +V C VCQ K EHQ P G++ L + EWKWD I+MDFV LP R
Sbjct: 932 YQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLP-VSR 990
Query: 284 GNDAIWVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRF 343
DAIWVIVDRLTKSAHFL I + LA+ Y+ EIV+LHGVP SIVSDRD +FTS F
Sbjct: 991 TKDAIWVIVDRLTKSAHFLAIRKTDGAVLLAKKYVSEIVELHGVPVSIVSDRDSKFTSAF 1050
Query: 344 WKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYN 403
W++ Q +G+K+++S+AYHPQTDGQSERTIQ+LED+LR+CVL++GG W HL L+EF YN
Sbjct: 1051 WRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDRGGHWADHLSLVEFAYN 1110
Query: 404 NSYHSSIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSR 463
NSY +SI MAPFEALYGR CR PLCW + GER + G V +TTE++++++ +K +Q R
Sbjct: 1111 NSYQASIRMAPFEALYGRPCRTPLCWTQVGERSIYGADYVLETTERIRVLKLNMKEAQDR 1170
Query: 464 QKTYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRV 523
Q++Y DKRR++LEF+ GD V+L++ + G R++ KLTP+++GP++I+ERVG VAYR+
Sbjct: 1171 QRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLTPRYMGPFRIVERVGPVAYRL 1230
Query: 524 GLPPHLSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGK 583
LP + H VFHVS L+K + V+ ++ N+T+E PVRI +R++K LR K
Sbjct: 1231 ELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKILEDLQPNMTLEARPVRILERRIKELRRK 1290
Query: 584 EIPLVRVVWT--GATGESLTWELESKMLVSYPELF 616
+IPL++V+W G T E TWE E++M S+ + F
Sbjct: 1291 KIPLIKVLWNCDGVTEE--TWEPEARMKASFKKWF 1323
>At4g07830 putative reverse transcriptase
Length = 611
Score = 541 bits (1394), Expect = e-154
Identities = 293/619 (47%), Positives = 400/619 (64%), Gaps = 53/619 (8%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ GKV+AY SRQL+ HE NYPTHDLE+AAV F+DHKSLKY+F
Sbjct: 1 MQRGKVIAYGSRQLKKHEGNYPTHDLEMAAV------------------FTDHKSLKYIF 42
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+R RRW++L+ DYD + YHPGKANVV DALSRK + A + E L
Sbjct: 43 TQLELNLRLRRWMKLVADYDLEIAYHPGKANVVTDALSRK--RVGAAPGQSVETLVIEIG 100
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+C + + LG+ +D ++ L ++ AQ+ D + L+A + E +++F G
Sbjct: 101 ALRLCAVAREP--LGLEAVDQTDLLSRVQLAQEKD----EGLIAASKAEGFEYQFAANGT 154
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
+ GR+C+P ++E+++ IL E+H S SIHPGATKMY DLK+ + W G+KRDV +V
Sbjct: 155 ILVHGRVCVPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWVEE 214
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSA 299
C VCQ K+EHQ ++ L +PEWKWD I+MDFV LP R DAIWVIVDRLTKSA
Sbjct: 215 CDVCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLP-VSRTKDAIWVIVDRLTKSA 273
Query: 300 HFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSS 359
HFL I + A LA+ Y+ EIVKLHGVP SIVSDRD +FTS FW++ Q +G+K+++S+
Sbjct: 274 HFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMST 333
Query: 360 AYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALY 419
AYHPQTDGQSERTIQ+LED+LR+CVL+ G W HL L+EF YNNSY +SIGMAPFE LY
Sbjct: 334 AYHPQTDGQSERTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMAPFEVLY 393
Query: 420 GRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQE 479
GR CR LCW + GER + G VQ+ TE++++++ +K +Q+RQ++Y DKRRK+LEF+
Sbjct: 394 GRPCR-TLCWTQVGERSIYGADYVQEITERIRVLKLNMKEAQNRQRSYADKRRKELEFEV 452
Query: 480 GDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVS 539
GD V + G +S+ +RVG VA+R+ L + H VFHVS
Sbjct: 453 GDSV-------SQDGHVARSE-------------QRVGPVAFRLELSDVMRAFHKVFHVS 492
Query: 540 QLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVV--WTGATG 597
L+K + V+ ++ N+T+E PVR+ +R++K LR K+IPL++V+ G T
Sbjct: 493 MLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLRNCDGVTE 552
Query: 598 ESLTWELESKMLVSYPELF 616
E TWE E+++ + + F
Sbjct: 553 E--TWEPEARLKARFKKWF 569
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 536 bits (1380), Expect = e-152
Identities = 292/619 (47%), Positives = 393/619 (63%), Gaps = 74/619 (11%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ GKV+AYASRQLR HE NYPTHDLE+AAV+F LKIWR YLYG +VF+DHKSLKY+F
Sbjct: 778 MQRGKVIAYASRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIF 837
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+RQR+W+EL+ DYD + YHPGKANVVADALS K + A + E L
Sbjct: 838 TQPELNLRQRQWMELVADYDLEIAYHPGKANVVADALSHK--RVGAAPGQSVEALVSEIG 895
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+C + + LG+ +D ++ L ++ AQ+ D + L+A + E S
Sbjct: 896 ALRLCAVARE--PLGLEAVDRADLLTRVRLAQEKD----EGLIAAYKAEGS--------- 940
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
E++ + E DV Q V
Sbjct: 941 ------------EDVANWVAE------------------------------CDVCQLV-- 956
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSA 299
K EHQ P GM+ L +PEWKWD I++DFV LP R DAIWVIVDRLTKSA
Sbjct: 957 -------KAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLP-VSRTKDAIWVIVDRLTKSA 1008
Query: 300 HFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSS 359
HFL I + A LA+ Y+ EIVKLHGVP SIVSDRD +FTS FW++ Q +G+K+++S+
Sbjct: 1009 HFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMST 1068
Query: 360 AYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALY 419
AYHPQT GQSERTIQ+LED+LR+CVL+ GG W HL L+EF YNNSY +SIGMAPFEALY
Sbjct: 1069 AYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASIGMAPFEALY 1128
Query: 420 GRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQE 479
R CR PLC + GER + G VQ+TTE++++++ +K +Q RQ++Y DKRR++LEF+
Sbjct: 1129 ERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKRRRELEFEV 1188
Query: 480 GDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVS 539
GD V+L++ + G R++ KL+P+++GP++I+ERVG VAYR+ LP + H VFHVS
Sbjct: 1189 GDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVS 1248
Query: 540 QLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVW--TGATG 597
L+K + V+ ++ N+T+E PVR+ +R++K LR K+IPL++V+W G T
Sbjct: 1249 MLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLWDCDGVTK 1308
Query: 598 ESLTWELESKMLVSYPELF 616
E TWE E++M + + F
Sbjct: 1309 E--TWEPEARMKARFKKWF 1325
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 493 bits (1268), Expect = e-139
Identities = 268/615 (43%), Positives = 367/615 (59%), Gaps = 109/615 (17%)
Query: 4 GKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQK 63
GKV+ YASRQLR HE NYPTHDLE+AA++F LKIW YLYG + +VF+DHKSLK +F Q
Sbjct: 722 GKVIVYASRQLRKHEGNYPTHDLEMAAIIFALKIWGSYLYGGKVQVFTDHKSLKDIFTQP 781
Query: 64 ELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRDMSL 123
ELN+RQRRW+EL+ DYD + YHPGK NVVADALSRK + + E +L
Sbjct: 782 ELNLRQRRWMELVADYDLEIAYHPGKTNVVADALSRKRVGAAPGQSVE----------AL 831
Query: 124 VCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGVLRFR 183
V E +G L++ +VAR+ + G +
Sbjct: 832 VSE-------IGALRL--------------------CVVAREPLKLEAVDRAANGTILVH 864
Query: 184 GRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYSCLVC 243
R+C+P +EE+++ IL E+H S SIHPGATKMY DLK+ + W G+KRDVA +V C VC
Sbjct: 865 ERVCLPKDEELRREILSEAHASMFSIHPGATKMYRDLKRHYQWVGMKRDVANWVTECDVC 924
Query: 244 QKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSAHFLP 303
Q K EHQ P G++ L + EWK T A LP
Sbjct: 925 QLVKAEHQVPGGLLQSLPISEWK----------------------------KTDGAAVLP 956
Query: 304 INISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHP 363
+ Y+ EIVKLHGVP SI+S RD +FTS FW++ Q +G+K+++S+AYHP
Sbjct: 957 -----------KKYVSEIVKLHGVPVSILSHRDSKFTSAFWRAFQVEMGTKVQMSTAYHP 1005
Query: 364 QTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRC 423
QTDGQSERTIQ+LED+L++CVL+ GG W HL L++F YNNSY +SIGMAPFEALYGR C
Sbjct: 1006 QTDGQSERTIQTLEDMLQMCVLDWGGHWADHLSLVKFAYNNSYQASIGMAPFEALYGRPC 1065
Query: 424 RIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQEGDHV 483
R LCW + GE+ + G VQ+TTE++++++ +K +Q RQ++Y DKRR++LEF+ G
Sbjct: 1066 RTLLCWTQVGEKSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKRRRELEFEVGT-- 1123
Query: 484 FLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQK 543
+I+ERVG VAYR+ LP + HNVFHVS L+K
Sbjct: 1124 ---------------------------EIVERVGPVAYRLELPDVMRAFHNVFHVSMLRK 1156
Query: 544 YVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVW--TGATGESLT 601
+ V+ ++ N+T+E PVR+ +R++K +R K+IP+++V+W G T E T
Sbjct: 1157 CLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKEVRRKKIPMIKVLWDCDGVTEE--T 1214
Query: 602 WELESKMLVSYPELF 616
WE E+++ + + F
Sbjct: 1215 WEPEARIKARFKKWF 1229
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 484 bits (1246), Expect = e-137
Identities = 224/423 (52%), Positives = 313/423 (73%), Gaps = 1/423 (0%)
Query: 170 SDFKFDDQGVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGL 229
++++ + G + GR+C+P++ +K+ IL E+H+S SIHPG+ KMY DLK+ + W G+
Sbjct: 527 TEYQTSNNGTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGM 586
Query: 230 KRDVAQFVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRG-NDAI 288
++DVA++V C CQ K EHQ P+G++ L + EWKWD I+MDFVT LP + ++A+
Sbjct: 587 RKDVARWVAKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAV 646
Query: 289 WVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQ 348
WV+VDRLTKSAHF+ I+ +AE YI EI++LHG+P SIVSDRD RFTS+FW + Q
Sbjct: 647 WVVVDRLTKSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQ 706
Query: 349 EALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHS 408
+ALG+++ LS+AYHPQTDGQSERTIQ+LED+LR CVL+ GG W+ +L LIEF YNNS+ +
Sbjct: 707 KALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQA 766
Query: 409 SIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYH 468
SIGM+P+EALYGR CR PLCW GER + GP IV +TTE+++ ++ K+K +Q RQK+Y
Sbjct: 767 SIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYA 826
Query: 469 DKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPH 528
+KRRK+LEFQ GD V+L+ G GR KKL+P+++GPY+++ERVG VAY++ LPP
Sbjct: 827 NKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPK 886
Query: 529 LSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLV 588
L+ HNVFHVSQL+KY+ D ++ +++N+TVE PVRI DR K RGK L+
Sbjct: 887 LNVFHNVFHVSQLRKYLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMSKGTRGKSRDLL 946
Query: 589 RVV 591
+V+
Sbjct: 947 KVL 949
Score = 77.4 bits (189), Expect = 2e-14
Identities = 35/42 (83%), Positives = 38/42 (90%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYL 42
MQ+G V+AYASRQLR HEKNYPTHDLE+AAVVF LKIWR YL
Sbjct: 466 MQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSYL 507
>At3g11970 hypothetical protein
Length = 1499
Score = 377 bits (968), Expect = e-104
Identities = 214/619 (34%), Positives = 341/619 (54%), Gaps = 33/619 (5%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQEG +AY SRQL+ + + ++ EL AV+F ++ WRHYL S F + +D +SLKYL
Sbjct: 912 MQEGHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLL 971
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
+Q+ Q++WL L ++D+ + Y GK NVVADALSR V E+L
Sbjct: 972 EQRLNTPIQQQWLPKLLEFDYEIQYRQGKENVVADALSR---------VEGSEVLH---- 1018
Query: 121 MSLVCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFD-DQGV 179
+ M ++ + L+ I+ D + D++ A + DS F Q +
Sbjct: 1019 -------------MAMTVVECDLLKDIQAGYANDSQLQDIITALQRDPDSKKYFSWSQNI 1065
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
LR + +I +P N+ IK IL H S + H G + +K +F+W G+ +D+ ++ S
Sbjct: 1066 LRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKGLFYWKGMIKDIQAYIRS 1125
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSA 299
C CQ+ K + G++ PL +P+ W +SMDF+ LP G I V+VDRL+K+A
Sbjct: 1126 CGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLP-VSGGKTVIMVVVDRLSKAA 1184
Query: 300 HFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSS 359
HF+ ++ + +A Y+ + KLHG P+SIVSDRD FTS FW+ G L+L+S
Sbjct: 1185 HFIALSHPYSALTVAHAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTS 1244
Query: 360 AYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALY 419
AYHPQ+DGQ+E + LE LR ++ W L L E+ YN +YHSS M PFE +Y
Sbjct: 1245 AYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVY 1304
Query: 420 GRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQE 479
G+ + L + +V + +Q+ + + ++ + +Q R K + D+ R + EF+
Sbjct: 1305 GQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEI 1364
Query: 480 GDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHV 538
GD+V++++ P ++ ++KL+PK+ GPY+I++R G VAY++ LP + S +H VFHV
Sbjct: 1365 GDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSY-SQVHPVFHV 1423
Query: 539 SQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGE 598
SQL+ V + S + V ++D E +P ++ +RK+ +GK + V V W+ E
Sbjct: 1424 SQLKVLVGNVSTTVHLPSV-MQD--VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLE 1480
Query: 599 SLTWELESKMLVSYPELFA 617
TWE + ++PE A
Sbjct: 1481 EATWEFLFDLQKTFPEFEA 1499
>At1g36590 hypothetical protein
Length = 1499
Score = 372 bits (956), Expect = e-103
Identities = 213/619 (34%), Positives = 341/619 (54%), Gaps = 33/619 (5%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQEG +AY SRQL+ + + ++ EL AV+F ++ WRHYL S F + +D +SLKYL
Sbjct: 912 MQEGHPLAYISRQLKGKQLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLL 971
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
+Q+ Q++WL L ++D+ + Y GK NVVADALSR V E+L
Sbjct: 972 EQRLNTPIQQQWLPKLLEFDYEIQYRQGKENVVADALSR---------VEGSEVLH---- 1018
Query: 121 MSLVCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFD-DQGV 179
+ M ++ + L+ I+ D + D++ A + DS F Q +
Sbjct: 1019 -------------MAMTVVECDLLKDIQAGYANDSQLQDIITALQRDPDSKKYFSWSQNI 1065
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
LR + +I +P N+ IK IL H S + H G + +K +F+ G+ +D+ ++ S
Sbjct: 1066 LRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKGLFYSKGMIKDIQAYIRS 1125
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSA 299
C CQ+ K + G++ PL +P+ W +SMDF+ LP G I V+VDRL+K+A
Sbjct: 1126 CGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLP-VSGGKTVIMVVVDRLSKAA 1184
Query: 300 HFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSS 359
HF+ ++ + +A+ Y+ + KLHG P+SIVSDRD FTS FW+ G L+L+S
Sbjct: 1185 HFIALSHPYSALTVAQAYLDNVFKLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTS 1244
Query: 360 AYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALY 419
AYHPQ+DGQ+E + LE LR ++ W L L E+ YN +YHSS M PFE +Y
Sbjct: 1245 AYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVY 1304
Query: 420 GRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDLEFQE 479
G+ + L + +V + +Q+ + + ++ + +Q R K + D+ R + EF+
Sbjct: 1305 GQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEI 1364
Query: 480 GDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHV 538
GD+V++++ P ++ ++KL+PK+ GPY+I++R G VAY++ LP + S +H VFHV
Sbjct: 1365 GDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSY-SQVHPVFHV 1423
Query: 539 SQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGE 598
SQL+ V + S + V ++D E +P ++ +RK+ +GK + V V W+ E
Sbjct: 1424 SQLKVLVGNVSTTVHLPSV-MQD--VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLE 1480
Query: 599 SLTWELESKMLVSYPELFA 617
TWE + ++PE A
Sbjct: 1481 EATWEFLFDLQKTFPEFEA 1499
>At1g35370 hypothetical protein
Length = 1447
Score = 336 bits (862), Expect = 2e-92
Identities = 199/609 (32%), Positives = 319/609 (51%), Gaps = 64/609 (10%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ+G +AY SRQL+ + + ++ EL A +F ++ WRHYL S F + +D +SLKYL
Sbjct: 883 MQKGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLL 942
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSR----KTLHMSAMMVREFELLE 116
+Q+ Q++WL L ++D+ + Y GK N+VADALSR + LHM+ +V
Sbjct: 943 EQRLNTPVQQQWLPKLLEFDYEIQYRQGKENLVADALSRVEGSEVLHMALSIV------- 995
Query: 117 QFRDMSLVCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFD- 175
+ +FL+ I+ A + D D++ A Q D+ +
Sbjct: 996 -----------------------ECDFLKEIQVAYESDGVLKDIISALQQHPDAKKHYSW 1032
Query: 176 DQGVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQ 235
Q +LR + +I +P++ EI +L+ H S + G + +K +F+W G+ +D+
Sbjct: 1033 SQDILRRKSKIVVPNDVEITNKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQA 1092
Query: 236 FVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRL 295
F+ SC CQ+ K ++ G++ PL +P+ W +SMDF+ LPN+ G I V+VDRL
Sbjct: 1093 FIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIWCDVSMDFIEGLPNSG-GKSVIMVVVDRL 1151
Query: 296 TKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKL 355
+K+AHF+ + + +A+ ++ + K HG P+SIVSDRD FTS FWK + G +L
Sbjct: 1152 SKAAHFVALAHPYSALTVAQAFLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVEL 1211
Query: 356 RLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPF 415
R+SSAYHPQ+DGQ+E + LE+ LR + W+ LPL E+ YN +YHSS M PF
Sbjct: 1212 RMSSAYHPQSDGQTEVVNRCLENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPF 1271
Query: 416 EALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKRRKDL 475
E +YG+ I L + +V + +Q+ + ++ + +Q R K + D+ R +
Sbjct: 1272 ELVYGQAPPIHLPYLPGKSKVAVVARSLQERENMLLFLKFHLMRAQHRMKQFADQHRTER 1331
Query: 476 EFQEGDHVFLRVTPMTGVGRALK-SKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHN 534
F GD V++++ P L+ ++KL+PK+ GPY+I+E+ G V + N
Sbjct: 1332 TFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKYFGPYKIIEKCGEVM-----------VGN 1380
Query: 535 VFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTG 594
V +QL +PD E P I +RK+ +G+ +V V W G
Sbjct: 1381 VTTSTQLPSVLPD----------------IFEKAPEYILERKLVKRQGRAATMVLVKWIG 1424
Query: 595 ATGESLTWE 603
E TW+
Sbjct: 1425 EPVEEATWK 1433
>At3g29490 hypothetical protein
Length = 438
Score = 309 bits (791), Expect = 3e-84
Identities = 165/390 (42%), Positives = 236/390 (60%), Gaps = 57/390 (14%)
Query: 229 LKRDVAQFVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAI 288
+KRDVA +V C VCQ K EHQ P GM+ L +PEW NT
Sbjct: 1 MKRDVANWVAECDVCQLVKAEHQVPGGMLQSLPIPEW--------------NT------- 39
Query: 289 WVIVDRLTKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQ 348
FL I + A LA+ Y+ EIVKLHGVP+
Sbjct: 40 ------------FLAIRKTDGAAVLAKKYVSEIVKLHGVPAE------------------ 69
Query: 349 EALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHS 408
+G+K+++S+ YHPQTDGQ ERTIQ+LED+LR+CVL+ GG W HL L+EF YNNSY +
Sbjct: 70 --MGTKVQMSTPYHPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQA 127
Query: 409 SIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYH 468
IGMAPFEALYGR CR PLCW + GER + G VQ+TTE++++++ +K +Q RQ +Y
Sbjct: 128 GIGMAPFEALYGRPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYA 187
Query: 469 DKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPH 528
DKRR++LEF+ GD V+L++ + G R++ KL+ +++GP++I+ERVG VAY + LP
Sbjct: 188 DKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDV 247
Query: 529 LSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLV 588
+ H VFHVS L+K + V+ ++ N+T+E VR+ +R++K L+ K+I L+
Sbjct: 248 MRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKISLI 307
Query: 589 RVVW--TGATGESLTWELESKMLVSYPELF 616
+V+W G T E TW+ E++M + + F
Sbjct: 308 KVLWDCDGVTEE--TWQPEARMKARFKKWF 335
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 274 bits (701), Expect = 9e-74
Identities = 161/363 (44%), Positives = 207/363 (56%), Gaps = 62/363 (17%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ GKV+AYASRQL HE NYPTHDLE+AAV+F LKIWR YLYG + +VF+DHKSLKY+F
Sbjct: 803 MQHGKVIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIF 862
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+RQRRW+EL+ DYD + YHPGKANVV DALSRK + A + + E+L
Sbjct: 863 TQPELNLRQRRWMELVADYDLEIAYHPGKANVVVDALSRK--RVGAALGQSVEVLVSEIG 920
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+C + + LG+ +D ++ L ++ AQK D+G+
Sbjct: 921 ALRLCAVAREP--LGLEAVDRADLLTRVRLAQK----------------------KDEGL 956
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
ATKMY DLK+ + W G+K DVA +V
Sbjct: 957 R--------------------------------ATKMYRDLKRYYQWVGMKMDVANWVAE 984
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSA 299
C VCQ K EHQ GM+ L +PEWKWD I+MD V L R DAIWVIVDRLTKSA
Sbjct: 985 CDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGL-RVSRTKDAIWVIVDRLTKSA 1043
Query: 300 HFLPINISFPVAQLAEIYIKEIVKLHGVPSSI--VSDRDPRFTSRFWKSLQEALGSKLRL 357
HFL I + A LA+ ++ EIVKLHGVP ++ DR + + + L+ +G ++ L
Sbjct: 1044 HFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYL 1103
Query: 358 SSA 360
A
Sbjct: 1104 KMA 1106
Score = 136 bits (342), Expect = 4e-32
Identities = 67/170 (39%), Positives = 110/170 (64%), Gaps = 4/170 (2%)
Query: 449 KVQMIQEKIKASQSRQKTYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIG 508
K+ + +K +Q RQ++Y DKRR++LEF+ GD V+L++ + G R++ KL+P+++G
Sbjct: 1067 KLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMG 1126
Query: 509 PYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETL 568
P++I+ERV VAYR+ LP + H VFHVS L+K + + ++ N+T+E
Sbjct: 1127 PFKIVERVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAKIPEDLQPNMTLEAR 1186
Query: 569 PVRIDDRKVKTLRGKEIPLVRVVW--TGATGESLTWELESKMLVSYPELF 616
PVR+ +R++K LR K+IPL++V+W G T E TWE E++M + + F
Sbjct: 1187 PVRVLERRIKELRQKKIPLIKVLWDCDGVTEE--TWEPEARMKARFKKWF 1234
>At4g03840 putative transposon protein
Length = 973
Score = 270 bits (689), Expect = 2e-72
Identities = 131/274 (47%), Positives = 182/274 (65%), Gaps = 19/274 (6%)
Query: 343 FWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTY 402
FWK+ Q+ALG+++ LS+AYHPQTDGQSERTIQ+LED+LR C L+ GG W+ +L L
Sbjct: 690 FWKAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACALDWGGNWEKYLRL----- 744
Query: 403 NNSYHSSIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQS 462
ALYGR CR PLCW GER + GP IV +TTE+++ ++ K+K +Q
Sbjct: 745 --------------ALYGRACRTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQD 790
Query: 463 RQKTYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYR 522
RQK+Y +KRRK+LEFQ D V+L+ G GR KKL+P+++GPY+++ERVG VAY+
Sbjct: 791 RQKSYANKRRKELEFQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYK 850
Query: 523 VGLPPHLSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRG 582
+ LPP L+ HNVFHVSQL+K + + ++ +++N+TVE PV+I DR K RG
Sbjct: 851 LDLPPKLNAFHNVFHVSQLRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTKGTRG 910
Query: 583 KEIPLVRVVWTGATGESLTWELESKMLVSYPELF 616
K L++V+W E TWE E+KM ++ E F
Sbjct: 911 KSRDLLKVLWNCGGREQYTWETENKMKANFSEWF 944
Score = 269 bits (688), Expect = 3e-72
Identities = 133/275 (48%), Positives = 186/275 (67%), Gaps = 7/275 (2%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ+G V+AYASRQLR HEKNYPTHDLE+AAVVF LKIWR YLYG++ ++++DHKSLKY+F
Sbjct: 421 MQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYIF 480
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+RQRRW+EL+ DYD + YH GKAN VADALSR+ + A + +L+
Sbjct: 481 TQPELNLRQRRWMELVADYDLDIAYHAGKANQVADALSRRRSEVEAER-SQVDLVNMMGT 539
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+ V S + LG+ D + L I+ AQ+ D + + ++++ + G
Sbjct: 540 LH-VNALSKEVEPLGLGAADQANLLSRIRLAQERDEEIKGWAL----NNKTEYQTSNNGT 594
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
+ GR+C+P+N +K+ IL E+H+S SIHPG+ K+Y DLK+ + W G+K+DVA++V
Sbjct: 595 IVVNGRVCVPNNRALKEEILREAHQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAK 654
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDF 274
C CQ K EHQ P+G++ L +PEWKWD I+MDF
Sbjct: 655 CPTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDF 689
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 269 bits (688), Expect = 3e-72
Identities = 133/275 (48%), Positives = 187/275 (67%), Gaps = 7/275 (2%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ+G V+AYASRQL HEKNYPTHDLE+AAVVF LKIWR YLYG++ ++++DHKSLKY+F
Sbjct: 421 MQKGSVIAYASRQLWKHEKNYPTHDLEMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYIF 480
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+RQRRW+EL+ DYD + YHPGKAN VADALSR+ + A + +L+
Sbjct: 481 IQPELNLRQRRWMELVADYDLDIAYHPGKANQVADALSRRRSEVEAER-SQVDLVNMMGT 539
Query: 121 MSLVCEWSPQSVKLGMLKID-SEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+ V S + LG+ D ++ L I+ AQ+ D + + ++++ + G
Sbjct: 540 LH-VNALSKEVESLGLGAADQADLLSRIRLAQERDEEIKGWAL----NNKTEYQTSNNGT 594
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYS 239
+ GR+C+P++ +K+ IL E+H+S SIHPG+ KMY DLK+ + W G+K+DVA++V
Sbjct: 595 IVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAK 654
Query: 240 CLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDF 274
C CQ K EHQ P+G++ L +PEWKWD I+MDF
Sbjct: 655 CPTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDF 689
Score = 236 bits (601), Expect = 4e-62
Identities = 114/229 (49%), Positives = 157/229 (67%), Gaps = 19/229 (8%)
Query: 346 SLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNS 405
+ Q+ALG+++ LS+AYHPQTDGQSERTIQ+LED+LR CVL+ GG W+ +L L
Sbjct: 690 AFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLTL-------- 741
Query: 406 YHSSIGMAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQK 465
ALYGR CR PLCW GER + GP IV +TTE+++ ++ K+K + RQK
Sbjct: 742 -----------ALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQK 790
Query: 466 TYHDKRRKDLEFQEGDHVFLRVTPMTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGL 525
+Y +KRRK+LEFQ GD V+L+ G GR KKL+P+++GPY+++ERVG VAY++ L
Sbjct: 791 SYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDL 850
Query: 526 PPHLSNLHNVFHVSQLQKYVPDPSHVIQSDDVQVRDNLTVETLPVRIDD 574
PP L+ HNVFHVSQL+K + D ++ +++N+TVE PVRI D
Sbjct: 851 PPKLNAFHNVFHVSQLRKCLSDQEESVEDVPPGLKENMTVEAWPVRIMD 899
>At4g07850 putative polyprotein
Length = 1138
Score = 212 bits (540), Expect = 4e-55
Identities = 112/305 (36%), Positives = 174/305 (56%), Gaps = 15/305 (4%)
Query: 136 MLKIDSEFLRSIKEAQKVD------VKFVDLLV-------ARDQTEDSDFKFDDQGVLRF 182
++ D E L+ +K QK++ V+F++ +D +G L +
Sbjct: 688 VIHTDHESLKHLKGQQKLNKRHARWVEFIETFPYVIKYKKGKDNVVADALSRRHEGFLFY 747
Query: 183 RGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQFVYSCLV 242
R+CIP N ++++ + E+H L H G +K ++ F W +KRDV + C
Sbjct: 748 DNRLCIP-NSSLRELFIREAHGGGLMGHFGVSKTLKVMQDHFHWPHMKRDVERMCERCTT 806
Query: 243 CQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRLTKSAHFL 302
C+++K + Q P G+ PL +P W+ ISMDFV LP T G D+I+V+VDR +K AHF+
Sbjct: 807 CKQAKAKSQ-PHGLCTPLPIPLHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFI 865
Query: 303 PINISFPVAQLAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYH 362
P + + +A ++ +E+V+LHG+P +IVSDRD +F S FWK+L LG+KL S+ H
Sbjct: 866 PCHKTDDAMHIANLFFREVVRLHGMPKTIVSDRDTKFLSYFWKTLWSKLGTKLLFSTTCH 925
Query: 363 PQTDGQSERTIQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRR 422
PQTDGQ+E ++L LLR + + TW+ LP +EF YN+S HS+ +PF+ +YG
Sbjct: 926 PQTDGQTEVVNRTLSTLLRALIKKNLKTWEDCLPHVEFAYNHSVHSATKFSPFQIVYGFN 985
Query: 423 CRIPL 427
PL
Sbjct: 986 PITPL 990
Score = 99.0 bits (245), Expect = 7e-21
Identities = 47/100 (47%), Positives = 68/100 (68%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ+ K +AY S +L NYPT+D EL A+V L+ W+HYL+ F + +DH+SLK+L
Sbjct: 641 MQDKKPIAYFSEKLGGATLNYPTYDKELYALVRALQTWQHYLWPKEFVIHTDHESLKHLK 700
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRK 100
Q++LN R RW+E ++ + + + Y GK NVVADALSR+
Sbjct: 701 GQQKLNKRHARWVEFIETFPYVIKYKKGKDNVVADALSRR 740
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 202 bits (513), Expect = 6e-52
Identities = 114/337 (33%), Positives = 178/337 (51%), Gaps = 36/337 (10%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQEG +A+ SRQL+ + + ++ EL AV+FV++ WRHYL S F + +D +SLKYL
Sbjct: 303 MQEGHPLAFISRQLKGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLL 362
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSR----KTLHMSAMMVREFELLE 116
+Q+ Q++WL L ++D+ + Y GK N+VADALSR + LHM+ +V
Sbjct: 363 EQRLNTPIQQQWLPKLLEFDYEIQYKQGKENLVADALSRVEGSEVLHMALSVV------- 415
Query: 117 QFRDMSLVCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFD- 175
+ + L+ I+ D ++ Q DS +
Sbjct: 416 -----------------------ECDLLKEIQAGYVTDGDIQGIITILQQQADSKKHYTW 452
Query: 176 DQGVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIFWWSGLKRDVAQ 235
QGVLR + +I +P+N IK IL H S + H G + +K +F+W + +D+
Sbjct: 453 SQGVLRRKNKIVVPNNSGIKDTILRWLHCSGMGGHSGKEVTHQRVKGLFYWKSMVKDIQA 512
Query: 236 FVYSCLVCQKSKVEHQKPAGMMVPLDVPEWKWDSISMDFVTSLPNTPRGNDAIWVIVDRL 295
F+ SC CQ+ K ++ G++ PL +P+ W +SMDF+ LP G I V+VDRL
Sbjct: 513 FIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWSDVSMDFIDGLP-LSNGKTVIMVVVDRL 571
Query: 296 TKSAHFLPINISFPVAQLAEIYIKEIVKLHGVPSSIV 332
+K+AHF+ + + +A+ Y+ + KLHG PSSIV
Sbjct: 572 SKAAHFIALAHPYSAMTVAQAYLDNVFKLHGCPSSIV 608
Score = 87.8 bits (216), Expect = 2e-17
Identities = 49/156 (31%), Positives = 91/156 (57%), Gaps = 5/156 (3%)
Query: 412 MAPFEALYGRRCRIPLCWFESGERVVLGPAIVQQTTEKVQMIQEKIKASQSRQKTYHDKR 471
M P+EA+YG+ + L + +V + +Q+ + ++ + +Q R K D+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 472 RKDLEFQEGDHVFLRVTPMTGVGRALKS-KKLTPKFIGPYQILERVGTVAYRVGLPPHLS 530
+ EF+ GD+VF+++ P ++S +KL+PK+ GPY++++R G VAY++ LP + S
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPAN-S 743
Query: 531 NLHNVFHVSQLQKY---VPDPSHVIQSDDVQVRDNL 563
+H VFHVSQL+ V +H+++ + +R+NL
Sbjct: 744 QVHPVFHVSQLRVLVGTVTTSTHLLRCYLMSLRENL 779
>At3g29480 hypothetical protein
Length = 718
Score = 178 bits (451), Expect = 9e-45
Identities = 102/225 (45%), Positives = 142/225 (62%), Gaps = 27/225 (12%)
Query: 1 MQEGKVVAYASRQLRVHEKNYPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLF 60
MQ GKV+AYASRQLR HE NYPTHDLE++AV F+DHKSLKY+
Sbjct: 520 MQRGKVIAYASRQLRKHEGNYPTHDLEMSAV------------------FTDHKSLKYII 561
Query: 61 DQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKTLHMSAMMVREFELLEQFRD 120
Q ELN+RQRRW+EL+ DYD + YH GKA+VVADALSRK + ++ E L+ + R
Sbjct: 562 TQPELNLRQRRWMELVADYDLEIAYHHGKASVVADALSRKRVGVAPGQSVE-ALVSEIRA 620
Query: 121 MSLVCEWSPQSVKLGMLKIDS-EFLRSIKEAQKVDVKFVDLLVARDQTEDSDFKFDDQGV 179
+ L C + + LG+ +D + L ++ AQ+ D + L+A + E S ++F G
Sbjct: 621 LRL-CAVARE--PLGLEAVDRVDLLSRVRLAQEKD----EGLIAASKAEGSKYQFAANGT 673
Query: 180 LRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLKKIF 224
+ GR+C+ ++EE+++ IL E+H S SIHPGATKMY DLK+ +
Sbjct: 674 ILVHGRVCVLNDEELRREILSEAHASMFSIHPGATKMYRDLKEYY 718
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 175 bits (444), Expect = 6e-44
Identities = 111/307 (36%), Positives = 168/307 (54%), Gaps = 11/307 (3%)
Query: 313 LAEIYIKEIVKLHGVPSSIVSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERT 372
+A ++ +VKLHG+P SIVSD DP F S FW+ + +KL +S+AYHPQTDGQ+E
Sbjct: 634 VAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTDGQTEVV 693
Query: 373 IQSLEDLLRICVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRIPLCWFES 432
+ +E LR V W S +P E+ YN ++H+S GM PF+ALYGR P+ +E
Sbjct: 694 NRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRP-PSPIPAYEL 752
Query: 433 GERVVLGPAIVQQTT--EKVQMIQEKIKASQSRQKTYHDKRRKDLEFQEGDHVFLRVTP- 489
G VV G Q E + +++ + + + K D R +D+ FQ GD V LR+ P
Sbjct: 753 GS-VVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLRIQPY 811
Query: 490 --MTGVGRALKSKKLTPKFIGPYQILERVGTVAYRVGLPPHLSNLHNVFHVSQLQKYVPD 547
T R+ S+KL+ +F GP+Q+ + G VAYR+ LP + +H VFHVS L+ +V D
Sbjct: 812 RQKTLFRRS--SQKLSHRFYGPFQVASKHGEVAYRLTLPEG-TRIHPVFHVSLLKPWVGD 868
Query: 548 PSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVRVVWTGATGESLTWELESK 607
+R+N ++ P + + + ++ K + + V W G E TWE +
Sbjct: 869 -GEPDMGQLPPLRNNGELKLQPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEEYDQ 927
Query: 608 MLVSYPE 614
+ S+PE
Sbjct: 928 LAASFPE 934
>At3g31490 hypothetical protein
Length = 285
Score = 168 bits (426), Expect = 7e-42
Identities = 92/225 (40%), Positives = 135/225 (59%), Gaps = 23/225 (10%)
Query: 42 LYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDFGLNYHPGKANVVADALSRKT 101
+YG + +VF+DHKSLKY+F Q+ELN+RQRRW+EL+ DYD + YHPGKANV +AL
Sbjct: 84 IYGGKVQVFTDHKSLKYMFTQQELNLRQRRWMELVVDYDLEIAYHPGKANVSVEAL---- 139
Query: 102 LHMSAMMVREFELLEQFRDMSLVCEWSPQSVKLGMLKIDSEFLRSIKEAQKVDVKFVDLL 161
V E L +C + + ++L ++ ++ L ++ AQ+ D + L
Sbjct: 140 -------VSEISALR-------LCAVAREPLELEVVG-RADLLSRVRLAQEKD----EGL 180
Query: 162 VARDQTEDSDFKFDDQGVLRFRGRICIPDNEEIKKMILEESHRSSLSIHPGATKMYHDLK 221
+A +TE S+ +F G + +IC P +E+++ IL E H S SI+P ATKMY DLK
Sbjct: 181 IAASKTEGSENQFAANGTILVHAQICGPKDEKLRWKILSEVHASMFSIYPRATKMYRDLK 240
Query: 222 KIFWWSGLKRDVAQFVYSCLVCQKSKVEHQKPAGMMVPLDVPEWK 266
+ + W G+KRDVA ++ C VCQ K EHQ P ++ L +PE K
Sbjct: 241 RYYQWVGMKRDVANWIAECDVCQLVKAEHQVPGALLQSLPIPEKK 285
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,939,445
Number of Sequences: 26719
Number of extensions: 599758
Number of successful extensions: 1646
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 1494
Number of HSP's gapped (non-prelim): 101
length of query: 617
length of database: 11,318,596
effective HSP length: 105
effective length of query: 512
effective length of database: 8,513,101
effective search space: 4358707712
effective search space used: 4358707712
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC138199.5


