

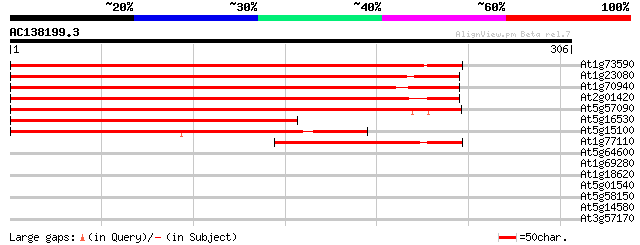
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.3 - phase: 0
(306 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g73590 auxin transporter splice variant b, putative 437 e-123
At1g23080 auxin transport protein (PIN7) 373 e-104
At1g70940 auxin transport like protein 368 e-102
At2g01420 auxin transporter splice variant b (PIN4) 360 e-100
At5g57090 root gravitropism control protein (PIN2) 359 1e-99
At5g16530 auxin efflux carrier -like protein 180 7e-46
At5g15100 auxin transport protein - like 173 9e-44
At1g77110 auxin transport protein (PIN6) 91 1e-18
At5g64600 auxin-independent growth promoter-like protein 31 0.73
At1g69280 hypothetical protein 30 1.3
At1g18620 hypothetical protein 30 1.3
At5g01540 receptor like protein kinase 30 2.1
At5g58150 receptor-like protein kinase 28 6.2
At5g14580 polynucleotide phosphorylase 28 6.2
At3g57170 unknown protein 28 6.2
>At1g73590 auxin transporter splice variant b, putative
Length = 622
Score = 437 bits (1123), Expect = e-123
Identities = 216/247 (87%), Positives = 232/247 (93%), Gaps = 1/247 (0%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT DFYHVMTAMVPLYVAMILAYGSVKWWKIF+PDQCSGINRFVALFAVPLLSFHFIA
Sbjct: 1 MITAADFYHVMTAMVPLYVAMILAYGSVKWWKIFTPDQCSGINRFVALFAVPLLSFHFIA 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+NNPY MNLRFLAAD+LQK+I+L LL +W S+ G L+WTITLFSLSTLPNTLVMGIPL
Sbjct: 61 ANNPYAMNLRFLAADSLQKVIVLSLLFLWCKLSRNGSLDWTITLFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
LKGMYG FSG LMVQIVVLQCIIWYTLMLF+FE+RGA+LLI EQFPDTAGSIVSIHVDSD
Sbjct: 121 LKGMYGNFSGDLMVQIVVLQCIIWYTLMLFLFEYRGAKLLISEQFPDTAGSIVSIHVDSD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
+MSLDGR PLET+AEIKEDGKLH+TVR+SNASRSDIYSRRSQGLS+ TPRPSNLTNAEIY
Sbjct: 181 IMSLDGRQPLETEAEIKEDGKLHVTVRRSNASRSDIYSRRSQGLSA-TPRPSNLTNAEIY 239
Query: 241 SLQSSRN 247
SLQSSRN
Sbjct: 240 SLQSSRN 246
>At1g23080 auxin transport protein (PIN7)
Length = 619
Score = 373 bits (957), Expect = e-104
Identities = 184/245 (75%), Positives = 213/245 (86%), Gaps = 3/245 (1%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V+TA++PLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITWHDLYTVLTAVIPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
SNNPY MNLRF+AADTLQKLI+L LL IW+NF++ G LEW+IT+FSLSTLPNTLVMGIPL
Sbjct: 61 SNNPYAMNLRFIAADTLQKLIMLTLLIIWANFTRSGSLEWSITIFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYGE+SGSLMVQIVVLQCIIWYTL+LF+FE+RGA++LI EQFP+T SIVS V+SD
Sbjct: 121 LIAMYGEYSGSLMVQIVVLQCIIWYTLLLFLFEYRGAKILIMEQFPETGASIVSFKVESD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
V+SLDG LETDA+I +DGKLH+TVRKSNASR Y G ++ TPRPSNLT AEIY
Sbjct: 181 VVSLDGHDFLETDAQIGDDGKLHVTVRKSNASRRSFY---GGGGTNMTPRPSNLTGAEIY 237
Query: 241 SLQSS 245
SL ++
Sbjct: 238 SLNTT 242
>At1g70940 auxin transport like protein
Length = 640
Score = 368 bits (945), Expect = e-102
Identities = 183/245 (74%), Positives = 213/245 (86%), Gaps = 6/245 (2%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MI+ D Y V+TA++PLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MISWHDLYTVLTAVIPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+NNPY MNLRF+AADTLQK+I+L LL +W+NF++ G LEW+IT+FSLSTLPNTLVMGIPL
Sbjct: 61 TNNPYAMNLRFIAADTLQKIIMLSLLVLWANFTRSGSLEWSITIFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYGE+SGSLMVQIVVLQCIIWYTL+LF+FEFRGA++LI EQFP+TA SIVS V+SD
Sbjct: 121 LIAMYGEYSGSLMVQIVVLQCIIWYTLLLFLFEFRGAKMLIMEQFPETAASIVSFKVESD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
V+SLDG LETDAEI +DGKLH+TVRKSNA SRRS + TPRPSNLT AEIY
Sbjct: 181 VVSLDGHDFLETDAEIGDDGKLHVTVRKSNA------SRRSFCGPNMTPRPSNLTGAEIY 234
Query: 241 SLQSS 245
SL ++
Sbjct: 235 SLSTT 239
>At2g01420 auxin transporter splice variant b (PIN4)
Length = 612
Score = 360 bits (924), Expect = e-100
Identities = 179/245 (73%), Positives = 206/245 (84%), Gaps = 9/245 (3%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V+TA+VPLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITWHDLYTVLTAVVPLYVAMILAYGSVQWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+N+PY MN RF+AADTLQK+I+L LLA+W+N +K G LEW IT+FSLSTLPNTLVMGIPL
Sbjct: 61 TNDPYAMNFRFVAADTLQKIIMLVLLALWANLTKNGSLEWMITIFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYG ++GSLMVQ+VVLQCIIWYTL+LF+FE+RGA+LLI EQFP+T SIVS V+SD
Sbjct: 121 LIAMYGTYAGSLMVQVVVLQCIIWYTLLLFLFEYRGAKLLIMEQFPETGASIVSFKVESD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
V+SLDG LETDAEI DGKLH+TVRKSNASR + TPRPSNLT AEIY
Sbjct: 181 VVSLDGHDFLETDAEIGNDGKLHVTVRKSNASRRSLMM---------TPRPSNLTGAEIY 231
Query: 241 SLQSS 245
SL S+
Sbjct: 232 SLSST 236
>At5g57090 root gravitropism control protein (PIN2)
Length = 647
Score = 359 bits (921), Expect = 1e-99
Identities = 183/251 (72%), Positives = 211/251 (83%), Gaps = 5/251 (1%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V+ AMVPLYVAMILAYGSV+WW IF+PDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITGKDMYDVLAAMVPLYVAMILAYGSVRWWGIFTPDQCSGINRFVAVFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
SN+PY MN FLAAD+LQK++IL L +W FS+RG LEW ITLFSLSTLPNTLVMGIPL
Sbjct: 61 SNDPYAMNYHFLAADSLQKVVILAALFLWQAFSRRGSLEWMITLFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L+ MYG+FSG+LMVQIVVLQ IIWYTLMLF+FEFRGA+LLI EQFP+TAGSI S VDSD
Sbjct: 121 LRAMYGDFSGNLMVQIVVLQSIIWYTLMLFLFEFRGAKLLISEQFPETAGSITSFRVDSD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYS---RRSQGLSSN--TPRPSNLT 235
V+SL+GR PL+TDAEI +DGKLH+ VR+S+A+ S I S GL+S+ TPR SNLT
Sbjct: 181 VISLNGREPLQTDAEIGDDGKLHVVVRRSSAASSMISSFNKSHGGGLNSSMITPRASNLT 240
Query: 236 NAEIYSLQSSR 246
EIYS+QSSR
Sbjct: 241 GVEIYSVQSSR 251
>At5g16530 auxin efflux carrier -like protein
Length = 351
Score = 180 bits (457), Expect = 7e-46
Identities = 86/157 (54%), Positives = 111/157 (69%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MI D Y V+ AMVPLYVA+IL YGSVKWW IF+ DQC INR V F +PL + F A
Sbjct: 1 MINCGDVYKVIEAMVPLYVALILGYGSVKWWHIFTRDQCDAINRLVCYFTLPLFTIEFTA 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+P+ MN RF+AAD L K+II+ +LA+W+ +S +G W+IT FSL TL N+LV+G+PL
Sbjct: 61 HVDPFNMNYRFIAADVLSKVIIVTVLALWAKYSNKGSYCWSITSFSLCTLTNSLVVGVPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGA 157
K MYG+ + L+VQ V Q I+W TL+LF+ EFR A
Sbjct: 121 AKAMYGQQAVDLVVQSSVFQAIVWLTLLLFVLEFRKA 157
>At5g15100 auxin transport protein - like
Length = 367
Score = 173 bits (439), Expect = 9e-44
Identities = 87/200 (43%), Positives = 131/200 (65%), Gaps = 10/200 (5%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MI+ D YHV++A VPLYV+M L + S + K+FSP+QC+GIN+FVA F++PLLSF I+
Sbjct: 1 MISWLDIYHVVSATVPLYVSMTLGFLSARHLKLFSPEQCAGINKFVAKFSIPLLSFQIIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNF-----SKRGCLEWTITLFSLSTLPNTLV 115
NNP+KM+ + + +D LQK +++ +LA+ F + G L W IT S+S LPNTL+
Sbjct: 61 ENNPFKMSPKLILSDILQKFLVVVVLAMVLRFWHPTGGRGGKLGWVITGLSISVLPNTLI 120
Query: 116 MGIPLLKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSI 175
+G+P+L +YG+ + S++ QIVVLQ +IWYT++LF+FE AR L P + S+
Sbjct: 121 LGMPILSAIYGDEAASILEQIVVLQSLIWYTILLFLFELNAARAL-----PSSGASLEHT 175
Query: 176 HVDSDVMSLDGRTPLETDAE 195
D + +++ E D E
Sbjct: 176 GNDQEEANIEDEPKEEEDEE 195
>At1g77110 auxin transport protein (PIN6)
Length = 429
Score = 90.5 bits (223), Expect = 1e-18
Identities = 51/104 (49%), Positives = 67/104 (64%), Gaps = 4/104 (3%)
Query: 145 YTLMLFMFEFRGARLLIFEQFP-DTAGSIVSIHVDSDVMSLDGRTPLETDAEIKEDGKLH 203
YTL+LF+FE R ARLLI +FP AGSI I VD DV+SLDG PL T+ E +G++
Sbjct: 4 YTLLLFLFELRAARLLIRAEFPGQAAGSIAKIQVDDDVISLDGMDPLRTETETDVNGRIR 63
Query: 204 ITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIYSLQSSRN 247
+ +R+S +S D S L TPR SNL+NAEI+S+ + N
Sbjct: 64 LRIRRSVSSVPDSVMSSSLCL---TPRASNLSNAEIFSVNTPNN 104
>At5g64600 auxin-independent growth promoter-like protein
Length = 539
Score = 31.2 bits (69), Expect = 0.73
Identities = 24/109 (22%), Positives = 49/109 (44%), Gaps = 12/109 (11%)
Query: 129 SGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHV--DSDVMSLDG 186
+ L + + L+C + Y + F + E+ AG +++H+ + D+++ G
Sbjct: 254 NNDLPIDVQRLRCRVLYRGLCFSPAIESLGQKLVERLKSRAGRYIALHLRYEKDMLAFTG 313
Query: 187 RTPLETDAEIKE-----DGKLHITVRKSNASRSDIYSRRSQGLSSNTPR 230
T TDAE +E + H ++ N++ +R +GL TP+
Sbjct: 314 CTYGLTDAESEELRVMRESTSHWKIKSINST-----EQREEGLCPLTPK 357
>At1g69280 hypothetical protein
Length = 278
Score = 30.4 bits (67), Expect = 1.3
Identities = 20/59 (33%), Positives = 32/59 (53%), Gaps = 8/59 (13%)
Query: 192 TDAEIKEDGKLHITVRKSNA------SRSDIYSRRSQGLSSNTPRPSNLTNAEIYSLQS 244
+D ++ED + IT R+S + S DI+ ++G SN S+L N E++SL S
Sbjct: 56 SDVRLEEDPE--ITTRRSTSLSLSPTSSEDIHEINAKGRKSNLSFDSSLHNKELFSLSS 112
>At1g18620 hypothetical protein
Length = 978
Score = 30.4 bits (67), Expect = 1.3
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 5/64 (7%)
Query: 182 MSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGL-----SSNTPRPSNLTN 236
MS RT +E I+ GK + + A S++ +S SS P P ++ N
Sbjct: 625 MSNKSRTKIEATLSIENGGKSPSVIEAAKAVVSNLIQNKSSPTFSEDGSSEHPSPVSVLN 684
Query: 237 AEIY 240
AEIY
Sbjct: 685 AEIY 688
>At5g01540 receptor like protein kinase
Length = 682
Score = 29.6 bits (65), Expect = 2.1
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 6/56 (10%)
Query: 190 LETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIYSLQSS 245
+E A IK DG L +T RKSN + + Y + + L+ N+ TN I S +S
Sbjct: 47 IEGAAMIKPDGLLRLTDRKSNVTGTAFYHKPVRLLNRNS------TNVTIRSFSTS 96
>At5g58150 receptor-like protein kinase
Length = 785
Score = 28.1 bits (61), Expect = 6.2
Identities = 23/80 (28%), Positives = 39/80 (48%), Gaps = 7/80 (8%)
Query: 175 IHVDSDVMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYS---RRSQGLSSN---T 228
+H+ + + L G P T ++ + L ++ K + SD++S S LSSN
Sbjct: 68 LHISASGLDLSGSIPDNTIGKMSKLQTLDLSGNKITSLPSDLWSLSLLESLNLSSNRISE 127
Query: 229 PRPSNLTN-AEIYSLQSSRN 247
P PSN+ N +++L S N
Sbjct: 128 PLPSNIGNFMSLHTLDLSFN 147
>At5g14580 polynucleotide phosphorylase
Length = 991
Score = 28.1 bits (61), Expect = 6.2
Identities = 17/55 (30%), Positives = 31/55 (55%), Gaps = 6/55 (10%)
Query: 168 TAGSIVSIHVDSDVMSLDGRTPLETDAE----IKEDGKLHITVRKSNASRSDIYS 218
TA +++ + + +L+ P ET AE +K+DGKL K+N++ S++ S
Sbjct: 841 TASKLIATQKEEE--ALESIAPEETSAECGEILKQDGKLKSVSPKNNSTASNLVS 893
>At3g57170 unknown protein
Length = 560
Score = 28.1 bits (61), Expect = 6.2
Identities = 23/70 (32%), Positives = 35/70 (49%), Gaps = 3/70 (4%)
Query: 115 VMGIPLLKGMYGEFSGSL-MVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIV 173
+MG+P + E +G L MV + V+Q IW TL +FM F + + T G+ V
Sbjct: 257 LMGVPAGFKLNTELAGVLGMVSLNVIQ--IWSTLWVFMASFIFCLIRVIAILGITFGATV 314
Query: 174 SIHVDSDVMS 183
S DV++
Sbjct: 315 SAAFVIDVIT 324
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,781,214
Number of Sequences: 26719
Number of extensions: 212264
Number of successful extensions: 728
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 702
Number of HSP's gapped (non-prelim): 21
length of query: 306
length of database: 11,318,596
effective HSP length: 99
effective length of query: 207
effective length of database: 8,673,415
effective search space: 1795396905
effective search space used: 1795396905
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138199.3


