

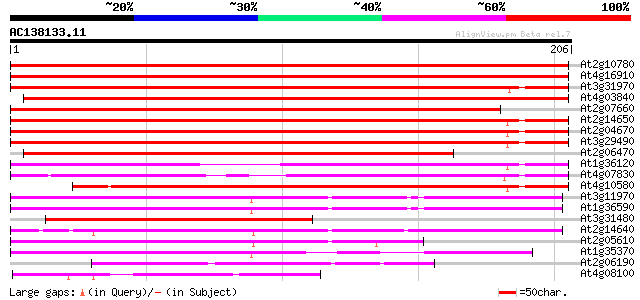
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.11 - phase: 0
(206 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 216 9e-57
At4g16910 retrotransposon like protein 206 7e-54
At3g31970 hypothetical protein 202 1e-52
At4g03840 putative transposon protein 198 2e-51
At2g07660 putative retroelement pol polyprotein 194 3e-50
At2g14650 putative retroelement pol polyprotein 194 4e-50
At2g04670 putative retroelement pol polyprotein 188 2e-48
At3g29490 hypothetical protein 185 2e-47
At2g06470 putative retroelement pol polyprotein 169 7e-43
At1g36120 putative reverse transcriptase gb|AAD22339.1 154 4e-38
At4g07830 putative reverse transcriptase 142 9e-35
At4g10580 putative reverse-transcriptase -like protein 138 2e-33
At3g11970 hypothetical protein 108 2e-24
At1g36590 hypothetical protein 108 2e-24
At3g31480 hypothetical protein 99 2e-21
At2g14640 putative retroelement pol polyprotein 87 5e-18
At2g05610 putative retroelement pol polyprotein 85 2e-17
At1g35370 hypothetical protein 62 2e-10
At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein 51 5e-07
At4g08100 putative polyprotein 46 2e-05
>At2g10780 pseudogene
Length = 1611
Score = 216 bits (549), Expect = 9e-57
Identities = 101/205 (49%), Positives = 141/205 (68%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
M+P+EALYGR CRTPLCW GE + GP +V ETTE++K ++ K+K +Q RQKSY +KR
Sbjct: 1378 MSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKR 1437
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
RK LEFQ GD V+L+ G GR +KL+P+++GPY++ ER+G VAY++ LPP L+
Sbjct: 1438 RKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNA 1497
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H+VFHVSQLRK ++D + +++N+TVE P+RI DR K RGK L++V+
Sbjct: 1498 FHNVFHVSQLRKCLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMTKGTRGKARDLLKVL 1557
Query: 181 WGGATGESLTWELESKMRDSYPELF 205
W E TWE E+KM+ ++PE F
Sbjct: 1558 WNCRGREEYTWETENKMKANFPEWF 1582
>At4g16910 retrotransposon like protein
Length = 687
Score = 206 bits (524), Expect = 7e-54
Identities = 97/205 (47%), Positives = 139/205 (67%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
M+P+EALYGR RTPLCW GE + GP +V ETT+K+K ++ K+K +Q RQKSY +KR
Sbjct: 462 MSPYEALYGRAGRTPLCWTPVGERRLFGPAVVDETTKKMKFLKIKLKEAQDRQKSYANKR 521
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
RK LEFQ GD V+L+ G GR +KL P+++GPY++ ER+G VAY++ LPP L
Sbjct: 522 RKELEFQVGDLVYLKAMTYKGAGRFTSRKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDA 581
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H+VFHVSQLRK +++ + +++N+TVE P+RI D+ K RGK + L++++
Sbjct: 582 FHNVFHVSQLRKCLSEQEESMEDVPPGLKENMTVEAWPVRIMDQMKKGTRGKSMDLLKIL 641
Query: 181 WGGATGESLTWELESKMRDSYPELF 205
W E TWE E+KM+ ++PE F
Sbjct: 642 WNCGGREEYTWETETKMKANFPEWF 666
>At3g31970 hypothetical protein
Length = 1329
Score = 202 bits (514), Expect = 1e-52
Identities = 98/207 (47%), Positives = 144/207 (69%), Gaps = 4/207 (1%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
MAPFEALYGR CRTPLCW + GE + G D V ETTE++++++ MK +Q RQ+SY DKR
Sbjct: 1119 MAPFEALYGRPCRTPLCWTQVGERSIYGADYVLETTERIRVLKLNMKEAQDRQRSYADKR 1178
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
R+ LEF+ GD V+L++ + G R++ KLTP+++GP++I ER+G VAYR+ LP +
Sbjct: 1179 RRELEFEVGDRVYLKMAMLRGPNRSISETKLTPRYMGPFRIVERVGPVAYRLELPDVMRA 1238
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H VFHVS LRK + V+ + ++ N+T+E P+RI +R++K LR K+IPL++V+
Sbjct: 1239 FHKVFHVSMLRKCLHKDDEVLAKILEDLQPNMTLEARPVRILERRIKELRRKKIPLIKVL 1298
Query: 181 WG--GATGESLTWELESKMRDSYPELF 205
W G T E TWE E++M+ S+ + F
Sbjct: 1299 WNCDGVTEE--TWEPEARMKASFKKWF 1323
>At4g03840 putative transposon protein
Length = 973
Score = 198 bits (503), Expect = 2e-51
Identities = 94/200 (47%), Positives = 134/200 (67%)
Query: 6 ALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKRRKALE 65
ALYGR CRTPLCW GE + GP +V ETTE++K ++ K+K +Q RQKSY +KRRK LE
Sbjct: 745 ALYGRACRTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQDRQKSYANKRRKELE 804
Query: 66 FQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVF 125
FQ D V+L+ G GR +KL+P+++GPY++ ER+G VAY++ LPP L+ H+VF
Sbjct: 805 FQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVF 864
Query: 126 HVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGAT 185
HVSQLRK +++ + +++N+TVE P++I DR K RGK L++V+W
Sbjct: 865 HVSQLRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTKGTRGKSRDLLKVLWNCGG 924
Query: 186 GESLTWELESKMRDSYPELF 205
E TWE E+KM+ ++ E F
Sbjct: 925 REQYTWETENKMKANFSEWF 944
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 194 bits (493), Expect = 3e-50
Identities = 91/180 (50%), Positives = 127/180 (70%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
M+P+EALYGR CRTPLCW GE + GP +V ETTE++K ++ K+K +Q RQKSY +KR
Sbjct: 770 MSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAQDRQKSYANKR 829
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
RK LEFQ GD V+L+ G GR +KL+P+++GPY++ ER+G VAY++ LPP L+
Sbjct: 830 RKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNV 889
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H+VFHVSQLRKY++D + +++N+TVE P+RI DR K RGK L++V+
Sbjct: 890 FHNVFHVSQLRKYLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMSKGTRGKSRDLLKVL 949
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 194 bits (492), Expect = 4e-50
Identities = 94/207 (45%), Positives = 142/207 (68%), Gaps = 4/207 (1%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
MAPFEALY R CRTPLC + GE + G D VQETTE++++++ MK +Q RQ+SY DKR
Sbjct: 1121 MAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKR 1180
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
R+ LEF+ GD V+L++ + G R++ KL+P+++GP++I ER+G VAYR+ LP +
Sbjct: 1181 RRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPVAYRLELPDVMRA 1240
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H VFHVS LRK + V+ + ++ N+T+E P+R+ +R++K LR K+IPL++V+
Sbjct: 1241 FHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVL 1300
Query: 181 W--GGATGESLTWELESKMRDSYPELF 205
W G T E TWE E++M+ + + F
Sbjct: 1301 WDCDGVTKE--TWEPEARMKARFKKWF 1325
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 188 bits (477), Expect = 2e-48
Identities = 92/207 (44%), Positives = 141/207 (67%), Gaps = 4/207 (1%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
MAPFEALYGR C TPL W + E + G D VQETTE++++++ MK +Q+RQ+SY DKR
Sbjct: 1201 MAPFEALYGRPCWTPLRWTQVEERSIYGADYVQETTERIRVLKLNMKEAQARQRSYADKR 1260
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
R+ LEF+ GD V+L++ + G R++ KL+P+++GP++I ER+G VAYR+ LP +
Sbjct: 1261 RRELEFEVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVERVGPVAYRLELPDVMRA 1320
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H VFHV LRK + V+ + ++ N+T+E P+R+ +R++K LR K+IPL++V+
Sbjct: 1321 FHKVFHVLMLRKCLHKDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVL 1380
Query: 181 W--GGATGESLTWELESKMRDSYPELF 205
W G T E TWE E++M+ + + F
Sbjct: 1381 WDCDGVTEE--TWEPEARMKARFKKWF 1405
>At3g29490 hypothetical protein
Length = 438
Score = 185 bits (469), Expect = 2e-47
Identities = 90/207 (43%), Positives = 139/207 (66%), Gaps = 4/207 (1%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
MAPFEALYGR CRTPLCW + GE + G D VQETTE++++++ MK +Q RQ SY DKR
Sbjct: 131 MAPFEALYGRPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYADKR 190
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
R+ LEF+ GD V+L++ + G R++ KL+ +++GP++I ER+G VAY + LP +
Sbjct: 191 RRELEFEVGDRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDVMRA 250
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H VFHVS LRK + V+ + ++ N+T+E +R+ +R++K L+ K+I L++V+
Sbjct: 251 FHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKISLIKVL 310
Query: 181 W--GGATGESLTWELESKMRDSYPELF 205
W G T E TW+ E++M+ + + F
Sbjct: 311 WDCDGVTEE--TWQPEARMKARFKKWF 335
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 169 bits (429), Expect = 7e-43
Identities = 79/158 (50%), Positives = 110/158 (69%)
Query: 6 ALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKRRKALE 65
ALYGR CRTPLCW GE + GP +V ETTE++K ++ K+K + RQKSY +KRRK LE
Sbjct: 742 ALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQKSYANKRRKELE 801
Query: 66 FQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVF 125
FQ GD V+L+ G GR +KL+P+++GPY++ ER+G VAY++ LPP L+ H+VF
Sbjct: 802 FQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVF 861
Query: 126 HVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDD 163
HVSQLRK ++D + +++N+TVE P+RI D
Sbjct: 862 HVSQLRKCLSDQEESVEDVPPGLKENMTVEAWPVRIMD 899
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 154 bits (388), Expect = 4e-38
Identities = 82/207 (39%), Positives = 123/207 (58%), Gaps = 33/207 (15%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
MAPFEALYGR CRT LCW + GE + G D VQETTE++++++ MK +Q RQ+SY DKR
Sbjct: 1054 MAPFEALYGRPCRTLLCWTQVGEKSIYGADYVQETTERIRVLKLNMKEAQDRQRSYADKR 1113
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
R+ LEF+ G +I ER+G VAYR+ LP +
Sbjct: 1114 RRELEFEVGT-----------------------------EIVERVGPVAYRLELPDVMRA 1144
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H+VFHVS LRK + V+ + ++ N+T+E P+R+ +R++K +R K+IP+++V+
Sbjct: 1145 FHNVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKEVRRKKIPMIKVL 1204
Query: 181 W--GGATGESLTWELESKMRDSYPELF 205
W G T E TWE E++++ + + F
Sbjct: 1205 WDCDGVTEE--TWEPEARIKARFKKWF 1229
>At4g07830 putative reverse transcriptase
Length = 611
Score = 142 bits (359), Expect = 9e-35
Identities = 82/207 (39%), Positives = 123/207 (58%), Gaps = 25/207 (12%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
MAPFE LYGR CRT LCW + GE + G D VQE TE++++++ MK +Q+RQ+SY DKR
Sbjct: 386 MAPFEVLYGRPCRT-LCWTQVGERSIYGADYVQEITERIRVLKLNMKEAQNRQRSYADKR 444
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSN 120
RK LEF+ GD V + G +S +R+G VA+R+ L +
Sbjct: 445 RKELEFEVGDSV-------SQDGHVARS-------------EQRVGPVAFRLELSDVMRA 484
Query: 121 LHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVV 180
H VFHVS LRK + V+ + ++ N+T+E P+R+ +R++K LR K+IPL++V+
Sbjct: 485 FHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVL 544
Query: 181 --WGGATGESLTWELESKMRDSYPELF 205
G T E TWE E++++ + + F
Sbjct: 545 RNCDGVTEE--TWEPEARLKARFKKWF 569
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 138 bits (347), Expect = 2e-33
Identities = 72/184 (39%), Positives = 115/184 (62%), Gaps = 5/184 (2%)
Query: 24 SVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVG 83
+ VL V E K+ + MK +Q RQ+SY DKRR+ LEF+ GD V+L++ + G
Sbjct: 1054 AAVLAKKFVSEIV-KLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPN 1112
Query: 84 RALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPR 143
R++ KL+P+++GP++I ER+ VAYR+ LP + H VFHVS LRK + + +
Sbjct: 1113 RSISETKLSPRYMGPFKIVERVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAK 1172
Query: 144 DDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVW--GGATGESLTWELESKMRDSY 201
++ N+T+E P+R+ +R++K LR K+IPL++V+W G T E TWE E++M+ +
Sbjct: 1173 IPEDLQPNMTLEARPVRVLERRIKELRQKKIPLIKVLWDCDGVTEE--TWEPEARMKARF 1230
Query: 202 PELF 205
+ F
Sbjct: 1231 KKWF 1234
>At3g11970 hypothetical protein
Length = 1499
Score = 108 bits (270), Expect = 2e-24
Identities = 64/204 (31%), Positives = 111/204 (54%), Gaps = 5/204 (2%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
M PFE +YG+ L + V + +QE + + ++ + +Q R K + D+
Sbjct: 1297 MTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRMKQFADQH 1356
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALK-SRKLTPKFIGPYQISERIGTVAYRVGLPPHLS 119
R EF+ GD+V++++ P ++ ++KL+PK+ GPY+I +R G VAY++ LP + S
Sbjct: 1357 RTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSY-S 1415
Query: 120 NLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRV 179
+H VFHVSQL+ V + S + V ++D E +P ++ +RK+ +GK + V V
Sbjct: 1416 QVHPVFHVSQLKVLVGNVSTTVHLPSV-MQD--VFEKVPEKVVERKMVNRQGKAVTKVLV 1472
Query: 180 VWGGATGESLTWELESKMRDSYPE 203
W E TWE ++ ++PE
Sbjct: 1473 KWSNEPLEEATWEFLFDLQKTFPE 1496
>At1g36590 hypothetical protein
Length = 1499
Score = 108 bits (270), Expect = 2e-24
Identities = 64/204 (31%), Positives = 111/204 (54%), Gaps = 5/204 (2%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
M PFE +YG+ L + V + +QE + + ++ + +Q R K + D+
Sbjct: 1297 MTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRMKQFADQH 1356
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALK-SRKLTPKFIGPYQISERIGTVAYRVGLPPHLS 119
R EF+ GD+V++++ P ++ ++KL+PK+ GPY+I +R G VAY++ LP + S
Sbjct: 1357 RTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSY-S 1415
Query: 120 NLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRV 179
+H VFHVSQL+ V + S + V ++D E +P ++ +RK+ +GK + V V
Sbjct: 1416 QVHPVFHVSQLKVLVGNVSTTVHLPSV-MQD--VFEKVPEKVVERKMVNRQGKAVTKVLV 1472
Query: 180 VWGGATGESLTWELESKMRDSYPE 203
W E TWE ++ ++PE
Sbjct: 1473 KWSNEPLEEATWEFLFDLQKTFPE 1496
>At3g31480 hypothetical protein
Length = 338
Score = 99.0 bits (245), Expect = 2e-21
Identities = 43/98 (43%), Positives = 68/98 (68%)
Query: 14 TPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKRRKALEFQEGDHVF 73
TPLCW + GE + G + V ETTE++++++ MK + RQ+SY D RR+ +EF+ GD V+
Sbjct: 111 TPLCWTQVGERSIYGANYVHETTERIQVLKLNMKEAHDRQRSYADMRRREVEFEVGDRVY 170
Query: 74 LRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYR 111
L++ + R + KL+PK++GP++I ER+G VAYR
Sbjct: 171 LKMDMLQSPKRFILETKLSPKYMGPFRIVERVGPVAYR 208
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 87.4 bits (215), Expect = 5e-18
Identities = 64/206 (31%), Positives = 109/206 (52%), Gaps = 7/206 (3%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGP--DLVQETTEKVKMIREKMKASQSRQKSYHD 58
M PF+ALYGR +P+ +E G SVV G + + E + +++ + + + K D
Sbjct: 733 MTPFQALYGRP-PSPIPAYELG-SVVCGELNEQMAARDELLAELKQHLVTANNCMKQQAD 790
Query: 59 KRRKALEFQEGDHVFLRVTPMTGVGRALKS-RKLTPKFIGPYQISERIGTVAYRVGLPPH 117
R + + FQ GD V LR+ P +S +KL+ +F GP+Q++ + G VAYR+ LP
Sbjct: 791 SRLRDVSFQVGDWVLLRIQPYRQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTLPEG 850
Query: 118 LSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLV 177
+ +H VFHVS L+ +V D + + +R+N ++ P + + + ++ K + +
Sbjct: 851 -TRIHPVFHVSLLKPWVGDGEPDMGQLP-PLRNNGELKLQPTAVLEVRWRSQDKKRVADL 908
Query: 178 RVVWGGATGESLTWELESKMRDSYPE 203
V W G E TWE ++ S+PE
Sbjct: 909 LVQWEGLHIEDATWEEYDQLAASFPE 934
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 85.1 bits (209), Expect = 2e-17
Identities = 51/156 (32%), Positives = 86/156 (54%), Gaps = 5/156 (3%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
M P+EA+YG+ L + V + +QE + ++ + +Q R K D+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALKS-RKLTPKFIGPYQISERIGTVAYRVGLPPHLS 119
EF+ GD+VF+++ P ++S +KL+PK+ GPY++ +R G VAY++ LP + S
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPAN-S 743
Query: 120 NLHDVFHVSQLRKY---VADPSHVIPRDDVQVRDNL 152
+H VFHVSQLR V +H++ + +R+NL
Sbjct: 744 QVHPVFHVSQLRVLVGTVTTSTHLLRCYLMSLRENL 779
>At1g35370 hypothetical protein
Length = 1447
Score = 62.0 bits (149), Expect = 2e-10
Identities = 49/193 (25%), Positives = 82/193 (42%), Gaps = 28/193 (14%)
Query: 1 MAPFEALYGRRCRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDKR 60
M PFE +YG+ L + V + +QE + ++ + +Q R K + D+
Sbjct: 1268 MTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQERENMLLFLKFHLMRAQHRMKQFADQH 1327
Query: 61 RKALEFQEGDHVFLRVTPMTGVGRALK-SRKLTPKFIGPYQISERIGTVAYRVGLPPHLS 119
R F GD V++++ P L+ ++KL+PK+ GPY+I E+ G V
Sbjct: 1328 RTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKYFGPYKIIEKCGEVM---------- 1377
Query: 120 NLHDVFHVSQLRKYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRV 179
+ +V +QL + D E P I +RK+ +G+ +V V
Sbjct: 1378 -VGNVTTSTQLPSVLPD----------------IFEKAPEYILERKLVKRQGRAATMVLV 1420
Query: 180 VWGGATGESLTWE 192
W G E TW+
Sbjct: 1421 KWIGEPVEEATWK 1433
>At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 280
Score = 50.8 bits (120), Expect = 5e-07
Identities = 34/126 (26%), Positives = 65/126 (50%), Gaps = 4/126 (3%)
Query: 31 LVQETTEKVKMIREKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGRALKSRK 90
+ +E ++++ K++A+ + K DKRR+ F+EGD V V G K
Sbjct: 154 MAEEILVVKEVVKAKLEATGKKNKVAADKRRRFKVFKEGDDVM--VLLRKGRFAVGTYNK 211
Query: 91 LTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPRDDVQVRD 150
+ P+ GP+++ +I AY V LP + N+ + F+V+ + +Y AD + P ++++
Sbjct: 212 VKPRKYGPFKVLRKINDNAYVVALPKSM-NISNTFNVADIHEYHAD-GVLYPEENLRTSS 269
Query: 151 NLTVET 156
+ ET
Sbjct: 270 SEVEET 275
>At4g08100 putative polyprotein
Length = 1054
Score = 45.8 bits (107), Expect = 2e-05
Identities = 36/119 (30%), Positives = 59/119 (49%), Gaps = 16/119 (13%)
Query: 2 APFEALYGRRCRTPLCWFE---SGESVVLGP---DLVQETTEKVKMIREKMKASQSRQKS 55
+PFE +YG + +PL S S + G DLVQ+ ++A + K
Sbjct: 857 SPFEIVYGFKPTSPLDLIPLPLSERSSLDGKKKDDLVQQVLN--------LEARTKQYKK 908
Query: 56 YHDKRRKALEFQEGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGL 114
Y +K RK + F EGD V++ + ++S KL P+ GP+++ +RI AY++ L
Sbjct: 909 YANKGRKEVIFNEGDQVWVHLRKKRFP--EVRSSKLMPRIDGPFKVLKRINNNAYKLDL 965
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,522,395
Number of Sequences: 26719
Number of extensions: 173306
Number of successful extensions: 540
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 486
Number of HSP's gapped (non-prelim): 44
length of query: 206
length of database: 11,318,596
effective HSP length: 95
effective length of query: 111
effective length of database: 8,780,291
effective search space: 974612301
effective search space used: 974612301
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC138133.11


