

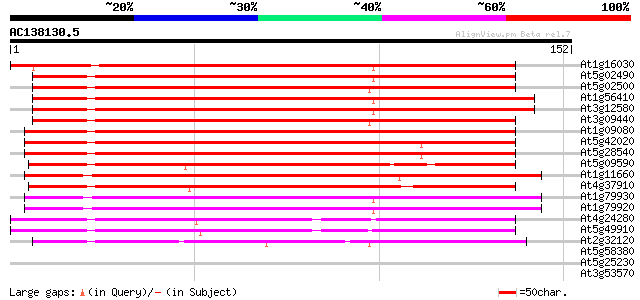
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.5 + phase: 0 /pseudo
(152 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g16030 unknown protein 189 4e-49
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 183 3e-47
At5g02500 dnaK-type molecular chaperone hsc70.1 182 5e-47
At1g56410 hypothetical protein 181 1e-46
At3g12580 putative protein 179 4e-46
At3g09440 heat-shock protein (At-hsc70-3) 178 9e-46
At1g09080 putative luminal binding protein 169 4e-43
At5g42020 luminal binding protein (dbj|BAA13948.1) 159 4e-40
At5g28540 luminal binding protein 159 4e-40
At5g09590 heat shock protein 70 (Hsc70-5) 106 6e-24
At1g11660 heat-shock protein like 105 9e-24
At4g37910 heat shock protein 70 like protein 103 3e-23
At1g79930 putative heat-shock protein 100 3e-22
At1g79920 putative heat-shock protein (At1g79920) 99 9e-22
At4g24280 hsp 70-like protein 91 2e-19
At5g49910 heat shock protein 70 (Hsc70-7) 89 9e-19
At2g32120 70kD heat shock protein 57 3e-09
At5g58380 SOS2-like protein kinase PKS2 29 1.1
At5g25230 translation Elongation Factor 2 - like protein 28 1.9
At3g53570 protein kinase (AME2/AFC1) 28 2.5
>At1g16030 unknown protein
Length = 646
Score = 189 bits (481), Expect = 4e-49
Identities = 96/140 (68%), Positives = 114/140 (80%), Gaps = 5/140 (3%)
Query: 1 MMTKS--VVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAK 58
M TKS +GIDLGTTYSC+ VW+ N RVEII NDQGNRTTPS+V+FT +RLIGDAAK
Sbjct: 1 MATKSEKAIGIDLGTTYSCVGVWM--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK 58
Query: 59 GQATSDPENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFC 117
Q +P+NTVFDAKRLIGRKFSD VQ+DI+ WPF+V+ G +KPMI+V YK +EK+F
Sbjct: 59 NQVALNPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQFS 118
Query: 118 PEEISSMILTKMREVAEKFL 137
PEEISSM+L KM+EVAE FL
Sbjct: 119 PEEISSMVLVKMKEVAEAFL 138
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 183 bits (465), Expect = 3e-47
Identities = 90/132 (68%), Positives = 109/132 (82%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPV 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+D LWPF +I G +KPMI+V+YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQSDRQLWPFTIISGTAEKPMIVVEYKGEEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFL 137
L KMRE+AE FL
Sbjct: 128 LIKMREIAEAFL 139
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 182 bits (463), Expect = 5e-47
Identities = 90/132 (68%), Positives = 108/132 (81%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPV 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRV-IGVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSDS VQ+D+ LWPF++ G DKPMI V+YKG+EK F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPADKPMIYVEYKGEEKEFAAEEISSMV 127
Query: 126 LTKMREVAEKFL 137
L KMRE+AE +L
Sbjct: 128 LIKMREIAEAYL 139
>At1g56410 hypothetical protein
Length = 617
Score = 181 bits (460), Expect = 1e-46
Identities = 91/137 (66%), Positives = 109/137 (79%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPV 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+FSD+ VQ+D+ WPF+V G DKPMI V YKG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRFSDASVQSDMKFWPFKVTPGQADKPMIFVNYKGEEKQFAAEEISSMV 127
Query: 126 LTKMREVAEKFLMSPRK 142
L KMRE+AE +L S K
Sbjct: 128 LIKMREIAEAYLGSSIK 144
>At3g12580 putative protein
Length = 650
Score = 179 bits (455), Expect = 4e-46
Identities = 90/137 (65%), Positives = 109/137 (78%), Gaps = 3/137 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPT 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR++SD VQ D WPF+V+ G +KPMI+V +KG+EK+F EEISSM+
Sbjct: 68 NTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQFSAEEISSMV 127
Query: 126 LTKMREVAEKFLMSPRK 142
L KMRE+AE FL SP K
Sbjct: 128 LIKMREIAEAFLGSPVK 144
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 178 bits (452), Expect = 9e-46
Identities = 89/132 (67%), Positives = 107/132 (80%), Gaps = 3/132 (2%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GIDLGTTYSC+ VW ++ RVEII NDQGNRTTPS+V+FT +RLIGDAAK Q +P
Sbjct: 10 IGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPI 67
Query: 67 NTVFDAKRLIGRKFSDSVVQNDIMLWPFRV-IGVDDKPMIIVKYKGQEKRFCPEEISSMI 125
NTVFDAKRLIGR+F+DS VQ+DI LWPF + G +KPMI+V YKG++K F EEISSMI
Sbjct: 68 NTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKSGPAEKPMIVVNYKGEDKEFSAEEISSMI 127
Query: 126 LTKMREVAEKFL 137
L KMRE+AE +L
Sbjct: 128 LIKMREIAEAYL 139
>At1g09080 putative luminal binding protein
Length = 678
Score = 169 bits (429), Expect = 4e-43
Identities = 85/133 (63%), Positives = 102/133 (75%), Gaps = 2/133 (1%)
Query: 5 SVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSD 64
+V+GIDLGTTYSC+ V+ N+ VEII NDQGNR TPS+V+FT +RLIG+AAK QA +
Sbjct: 51 TVIGIDLGTTYSCVGVY--HNKHVEIIANDQGNRITPSWVAFTDTERLIGEAAKNQAAKN 108
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYKGQEKRFCPEEISSM 124
PE T+FD KRLIGRKF D VQ DI P++V+ D KP I VK KG+EK F PEEIS+M
Sbjct: 109 PERTIFDPKRLIGRKFDDPDVQRDIKFLPYKVVNKDGKPYIQVKVKGEEKLFSPEEISAM 168
Query: 125 ILTKMREVAEKFL 137
ILTKM+E AE FL
Sbjct: 169 ILTKMKETAEAFL 181
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 159 bits (403), Expect = 4e-40
Identities = 84/134 (62%), Positives = 100/134 (73%), Gaps = 3/134 (2%)
Query: 5 SVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSD 64
SV+GIDLGTTYSC+ V+ +N VEII NDQGNR TPS+V FT +RLIG+AAK QA +
Sbjct: 36 SVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAVN 93
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYK-GQEKRFCPEEISS 123
PE TVFD KRLIGRKF D VQ D L P++++ D KP I VK K G+ K F PEEIS+
Sbjct: 94 PERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNKDGKPYIQVKIKDGETKVFSPEEISA 153
Query: 124 MILTKMREVAEKFL 137
MILTKM+E AE +L
Sbjct: 154 MILTKMKETAEAYL 167
>At5g28540 luminal binding protein
Length = 669
Score = 159 bits (403), Expect = 4e-40
Identities = 84/134 (62%), Positives = 100/134 (73%), Gaps = 3/134 (2%)
Query: 5 SVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSD 64
SV+GIDLGTTYSC+ V+ +N VEII NDQGNR TPS+V FT +RLIG+AAK QA +
Sbjct: 36 SVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAVN 93
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYK-GQEKRFCPEEISS 123
PE TVFD KRLIGRKF D VQ D L P++++ D KP I VK K G+ K F PEEIS+
Sbjct: 94 PERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNKDGKPYIQVKIKDGETKVFSPEEISA 153
Query: 124 MILTKMREVAEKFL 137
MILTKM+E AE +L
Sbjct: 154 MILTKMKETAEAYL 167
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 106 bits (264), Expect = 6e-24
Identities = 60/133 (45%), Positives = 86/133 (64%), Gaps = 6/133 (4%)
Query: 6 VVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSF-TKDQRLIGDAAKGQATSD 64
V+GIDLGTT SC+AV E + ++I N +G RTTPS V+F TK + L+G AK QA ++
Sbjct: 59 VIGIDLGTTNSCVAVM--EGKNPKVIENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTN 116
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYKGQEKRFCPEEISSM 124
P NTV KRLIGRKF D Q ++ + P++++ + V+ GQ+ + P +I +
Sbjct: 117 PTNTVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGD-AWVEANGQQ--YSPSQIGAF 173
Query: 125 ILTKMREVAEKFL 137
ILTKM+E AE +L
Sbjct: 174 ILTKMKETAEAYL 186
>At1g11660 heat-shock protein like
Length = 773
Score = 105 bits (262), Expect = 9e-24
Identities = 57/141 (40%), Positives = 87/141 (61%), Gaps = 3/141 (2%)
Query: 5 SVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSD 64
SVVG D+G IAV + R ++++ ND+ NR P+ VSF + QR +G AA AT
Sbjct: 2 SVVGFDVGNENCVIAV--AKQRGIDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMH 59
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPM-IIVKYKGQEKRFCPEEISS 123
P++T+ KRLIGRKF + VQND+ L+PF D + I ++Y G+ + F P +I
Sbjct: 60 PKSTISQLKRLIGRKFREPDVQNDLRLFPFETSEDSDGGIQIRLRYMGEIQSFSPVQILG 119
Query: 124 MILTKMREVAEKFLMSPRKEC 144
M+L+ ++++AEK L +P +C
Sbjct: 120 MLLSHLKQIAEKSLKTPVSDC 140
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 103 bits (258), Expect = 3e-23
Identities = 53/133 (39%), Positives = 85/133 (63%), Gaps = 6/133 (4%)
Query: 6 VVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFT-KDQRLIGDAAKGQATSD 64
V+GIDLGTT SC++V E + +I N +G+RTTPS V+ K + L+G AK QA ++
Sbjct: 54 VIGIDLGTTNSCVSVM--EGKTARVIENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTN 111
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYKGQEKRFCPEEISSM 124
P NT+F +KRLIGR+F D Q ++ + P++++ + + + ++F P +I +
Sbjct: 112 PTNTIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV---EANGQKFSPSQIGAN 168
Query: 125 ILTKMREVAEKFL 137
+LTKM+E AE +L
Sbjct: 169 VLTKMKETAEAYL 181
>At1g79930 putative heat-shock protein
Length = 831
Score = 100 bits (249), Expect = 3e-22
Identities = 54/141 (38%), Positives = 80/141 (56%), Gaps = 3/141 (2%)
Query: 5 SVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSD 64
SVVG D G +AV R ++++ ND+ NR TP+ V F QR IG A +
Sbjct: 2 SVVGFDFGNENCLVAV--ARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMN 59
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISS 123
P+N++ KRLIGR+FSD +Q DI PF V G D P+I Y G+++ F P ++
Sbjct: 60 PKNSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEKRAFTPTQVMG 119
Query: 124 MILTKMREVAEKFLMSPRKEC 144
M+L+ ++ +AEK L + +C
Sbjct: 120 MMLSNLKGIAEKNLNTAVVDC 140
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 99.0 bits (245), Expect = 9e-22
Identities = 54/141 (38%), Positives = 79/141 (55%), Gaps = 3/141 (2%)
Query: 5 SVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSD 64
SVVG D G +AV R ++++ ND+ NR TP+ V F QR IG A +
Sbjct: 2 SVVGFDFGNENCLVAV--ARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMN 59
Query: 65 PENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVI-GVDDKPMIIVKYKGQEKRFCPEEISS 123
P+N++ KRLIGR+FSD +Q DI PF V G D P+I Y G+ + F P ++
Sbjct: 60 PKNSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEIRAFTPTQVMG 119
Query: 124 MILTKMREVAEKFLMSPRKEC 144
M+L+ ++ +AEK L + +C
Sbjct: 120 MMLSNLKGIAEKNLNTAVVDC 140
>At4g24280 hsp 70-like protein
Length = 718
Score = 91.3 bits (225), Expect = 2e-19
Identities = 56/138 (40%), Positives = 82/138 (58%), Gaps = 6/138 (4%)
Query: 1 MMTKSVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKD-QRLIGDAAKG 59
++ + VVGIDLGTT S +A E + I+ N +G RTTPS V++TK RL+G AK
Sbjct: 75 VVNEKVVGIDLGTTNSAVAAM--EGGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKR 132
Query: 60 QATSDPENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYKGQEKRFCPE 119
QA +PENT F KR IGRK ++ V + +RV+ D+ + ++ K+F E
Sbjct: 133 QAVVNPENTFFSVKRFIGRKMNE--VDEESKQVSYRVVR-DENNNVKLECPAINKQFAAE 189
Query: 120 EISSMILTKMREVAEKFL 137
EIS+ +L K+ + A +FL
Sbjct: 190 EISAQVLRKLVDDASRFL 207
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 89.0 bits (219), Expect = 9e-19
Identities = 56/138 (40%), Positives = 82/138 (58%), Gaps = 6/138 (4%)
Query: 1 MMTKSVVGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQ-RLIGDAAKG 59
++ + VVGIDLGTT S +A E + I+ N +G RTTPS V++TK + RL+G AK
Sbjct: 75 VVNEKVVGIDLGTTNSAVAAM--EGGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKR 132
Query: 60 QATSDPENTVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYKGQEKRFCPE 119
QA +PENT F KR IGR+ ++ V + +RVI D+ + + K+F E
Sbjct: 133 QAVVNPENTFFSVKRFIGRRMNE--VAEESKQVSYRVI-KDENGNVKLDCPAIGKQFAAE 189
Query: 120 EISSMILTKMREVAEKFL 137
EIS+ +L K+ + A +FL
Sbjct: 190 EISAQVLRKLVDDASRFL 207
>At2g32120 70kD heat shock protein
Length = 563
Score = 57.4 bits (137), Expect = 3e-09
Identities = 45/139 (32%), Positives = 69/139 (49%), Gaps = 9/139 (6%)
Query: 7 VGIDLGTTYSCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPE 66
+GID+GT+ IAVW +V I+ N + + SFV+F KD+ G + A
Sbjct: 31 LGIDIGTSQCSIAVW--NGSQVHILRNTRNQKLIKSFVTF-KDEVPAGGVSNQLAHEQEM 87
Query: 67 NT---VFDAKRLIGRKFSDSVVQNDIMLWPFRV--IGVDDKPMIIVKYKGQEKRFCPEEI 121
T +F+ KRL+GR +D VV L PF V + + +P I + PEE+
Sbjct: 88 LTGAAIFNMKRLVGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNAWRSTTPEEV 146
Query: 122 SSMILTKMREVAEKFLMSP 140
++ L ++R +AE L P
Sbjct: 147 LAIFLVELRLMAEAQLKRP 165
>At5g58380 SOS2-like protein kinase PKS2
Length = 479
Score = 28.9 bits (63), Expect = 1.1
Identities = 12/31 (38%), Positives = 23/31 (73%)
Query: 112 QEKRFCPEEISSMILTKMREVAEKFLMSPRK 142
+E RF ++ +S+I++K+ EVA++ +S RK
Sbjct: 351 RESRFTSQKPASVIISKLEEVAQRLKLSIRK 381
>At5g25230 translation Elongation Factor 2 - like protein
Length = 973
Score = 28.1 bits (61), Expect = 1.9
Identities = 25/76 (32%), Positives = 35/76 (45%), Gaps = 8/76 (10%)
Query: 68 TVFDAKRLIGRKFSDSVVQNDIMLWPFRVIGVDDKPMIIVKYKGQEKRFCPEEI----SS 123
T D ++G K DS+VQ W R + D+P+ VK+K + R PE +
Sbjct: 752 TEVDRNLMMGVK--DSIVQG--FQWGAREGPLCDEPIRNVKFKIVDARIAPEPLHRGSGQ 807
Query: 124 MILTKMREVAEKFLMS 139
MI T R FLM+
Sbjct: 808 MIPTARRVAYSAFLMA 823
>At3g53570 protein kinase (AME2/AFC1)
Length = 467
Score = 27.7 bits (60), Expect = 2.5
Identities = 23/72 (31%), Positives = 28/72 (37%), Gaps = 7/72 (9%)
Query: 16 SCIAVWLDENRRVEIIHNDQGNRTTPSFVSFTKDQRLIGDAAKGQATSDPENTVFDAKRL 75
S IA+ L+ R VE H R K RL DAA P TVF
Sbjct: 11 SSIAMILETQRNVEFPHRIVDKRPR-------KRPRLTWDAAPPLLPPPPPPTVFQPPLY 63
Query: 76 IGRKFSDSVVQN 87
G +F+ +V N
Sbjct: 64 YGPEFASGLVPN 75
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,395,238
Number of Sequences: 26719
Number of extensions: 131877
Number of successful extensions: 375
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 319
Number of HSP's gapped (non-prelim): 29
length of query: 152
length of database: 11,318,596
effective HSP length: 90
effective length of query: 62
effective length of database: 8,913,886
effective search space: 552660932
effective search space used: 552660932
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC138130.5


