

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138087.2 + phase: 0 /pseudo
(296 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
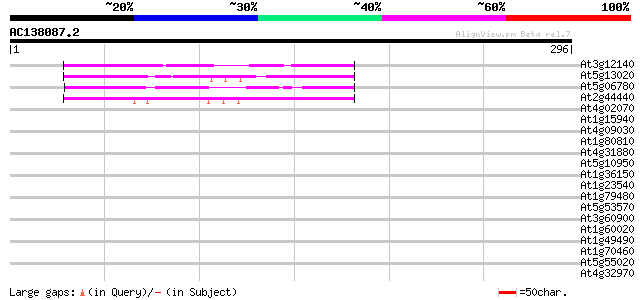
Score E
Sequences producing significant alignments: (bits) Value
At3g12140 unknown protein 136 2e-32
At5g13020 putative protein 135 3e-32
At5g06780 unknown protein 99 2e-21
At2g44440 hypothetical protein 78 5e-15
At4g02070 G/T DNA mismatch repair enzyme 37 0.013
At1g15940 T24D18.4 37 0.017
At4g09030 arabinogalactan-protein AGP10 32 0.31
At1g80810 unknown protein 31 0.91
At4g31880 unknown protein 30 1.2
At5g10950 putative protein 30 1.6
At1g36150 hypothetical protein 30 2.0
At1g23540 putative serine/threonine protein kinase 30 2.0
At1g79480 hypothetical protein 29 2.7
At5g53570 GTPase activator protein of Rab-like small GTPases-lik... 29 3.5
At3g60900 GPI-anchored protein (Fla10) 29 3.5
At1g60020 hypothetical protein 29 3.5
At1g49490 hypothetical protein 29 3.5
At1g70460 putative protein kinase 28 4.5
At5g55020 putative transcription factor MYB120 (MYB120) 28 5.9
At4g32970 putative protein 28 5.9
>At3g12140 unknown protein
Length = 327
Score = 136 bits (342), Expect = 2e-32
Identities = 79/154 (51%), Positives = 92/154 (59%), Gaps = 22/154 (14%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ESLITEL ELRVSD+EHRELL RVN D I R RDWR+ G Q T QP D+LPS
Sbjct: 32 ESLITELRKELRVSDDEHRELLSRVNKDDTIQRIRDWRQGGASQITRHATIQP-FDVLPS 90
Query: 89 PTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPAE 148
PT SA+ KK KT S PS+ G TG R F+N SS ++
Sbjct: 91 PTFSAARKKQKTFPSYNPSI------------------GATGNRSFNNRLVSSGISG--- 129
Query: 149 EAQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
+ LIG+KVWT+WPED+HFYEA+IT YN DE
Sbjct: 130 NESAEALIGRKVWTKWPEDNHFYEAIITQYNADE 163
>At5g13020 putative protein
Length = 414
Score = 135 bits (339), Expect = 3e-32
Identities = 84/178 (47%), Positives = 102/178 (57%), Gaps = 33/178 (18%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ESLITEL ELRVSDEEHRELL RVN+D +I R R+WR+ Q + Q VHD PS
Sbjct: 81 ESLITELRKELRVSDEEHRELLSRVNADEMIRRIREWRKANSLQS---SVPQLVHDA-PS 136
Query: 89 PTVSASNKKPKTSHSVP--------PSLGQSL-------------PGLSSVKP---VQCA 124
P VS S KK KTS S+ PSL S+ PG + KP +Q
Sbjct: 137 PAVSGSRKKQKTSQSIASLAMGPPSPSLHPSMQPSSSALRRGGPPPGPKTKKPKTSMQYP 196
Query: 125 STGPTGARHFSNHNSSSNLNAPAEEAQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
STG G + + N P E +DPL+G+KVWT+WP+D+ +YEAVITDYNP E
Sbjct: 197 STGIAG-----RPQAGALTNEPGESGSYDPLVGRKVWTKWPDDNQYYEAVITDYNPVE 249
>At5g06780 unknown protein
Length = 320
Score = 99.4 bits (246), Expect = 2e-21
Identities = 64/153 (41%), Positives = 84/153 (54%), Gaps = 30/153 (19%)
Query: 30 SLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPSP 89
+++T L ELR+SD+E+R+LL V++D +I R RD R G Q D+ PSP
Sbjct: 40 TVMTNLRKELRISDDENRQLLNNVHNDDLIKRIRDSRPRGGNQ----VVRHQSLDVHPSP 95
Query: 90 TVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPAEE 149
T SAS KK KT S P S G T ++ F+N S+N PAE
Sbjct: 96 TFSASRKKQKTFQSYP-------------------SIGSTRSKSFNNRVVSAN--EPAEA 134
Query: 150 AQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
LIG+KVWT+WPED+ FYEAV+T YN +E
Sbjct: 135 -----LIGRKVWTKWPEDNSFYEAVVTQYNANE 162
>At2g44440 hypothetical protein
Length = 343
Score = 78.2 bits (191), Expect = 5e-15
Identities = 65/192 (33%), Positives = 84/192 (42%), Gaps = 38/192 (19%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRD----------------WRETGIY- 71
ES+ITEL EL +S+EEHRELLGRVNSD I R R WR Y
Sbjct: 62 ESVITELRKELSLSNEEHRELLGRVNSDDTIRRIRQDLKSEHLSDEQGMETIWRNAAKYA 121
Query: 72 ----------QPPGCTTSQPVHDILPSPTVSASNKKPKTSHS-----VPPSLGQS----L 112
+P SQP PS A P S + VP G+
Sbjct: 122 QCCSTSMKTHKPNQPIPSQPFASSSPSFHPQADPTHPFASSTAKRGPVPIVKGKKHKPVF 181
Query: 113 PGLSSVK--PVQCASTGPTGARHFSNHNSSSNLNAPAEEAQFDPLIGKKVWTRWPEDSHF 170
PG SS K P + P G + ++ + P + +G++V T+WPED+ F
Sbjct: 182 PGSSSTKHAPYHPSDQPPRGQVMNRLPSVPASSSEPTNGIDPESFLGRRVRTKWPEDNTF 241
Query: 171 YEAVITDYNPDE 182
Y+A+IT YNP E
Sbjct: 242 YDAIITQYNPVE 253
>At4g02070 G/T DNA mismatch repair enzyme
Length = 1324
Score = 37.0 bits (84), Expect = 0.013
Identities = 32/115 (27%), Positives = 45/115 (38%), Gaps = 19/115 (16%)
Query: 87 PSPTVSASNKK-----------PKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFS 135
PSP+ S SNKK P S S P + P SS P + S GP
Sbjct: 37 PSPSPSLSNKKTPKSNNPNPKSPSPSPSPPKKTPKLNPNPSSNLPARSPSPGPDTPSPVQ 96
Query: 136 NHNSSSNL-----NAPAEEAQF---DPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
+ L +P + D ++GK+V WP D +Y+ +T Y+ E
Sbjct: 97 SKFKKPLLVIGQTPSPPQSVVITYGDEVVGKQVRVYWPLDKKWYDGSVTFYDKGE 151
>At1g15940 T24D18.4
Length = 1012
Score = 36.6 bits (83), Expect = 0.017
Identities = 28/93 (30%), Positives = 38/93 (40%), Gaps = 15/93 (16%)
Query: 86 LPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNA 145
+ +P +S K K S + P+ + K Q + P R + SN N
Sbjct: 539 MDTPIPQSSKSKKKDSRATTPA---------TKKSEQAPKSHPKMKR-IAGEEVESNTNE 588
Query: 146 PAEEAQFDPLIGKKVWTRWPEDSHFYEAVITDY 178
EE L+GK+V WP D FYE VI Y
Sbjct: 589 LGEE-----LVGKRVNVWWPLDKKFYEGVIKSY 616
>At4g09030 arabinogalactan-protein AGP10
Length = 127
Score = 32.3 bits (72), Expect = 0.31
Identities = 24/77 (31%), Positives = 35/77 (45%), Gaps = 11/77 (14%)
Query: 72 QPPGCTTSQPVHDILPSPTVSASNKKPKTSHSVPPSLGQSLPGLSS-------VKPVQCA 124
QPP T+ P I P+PT + S T+ V P G LP +S + P
Sbjct: 38 QPP--RTAAPTPSITPTPTPTPS--ATPTAAPVSPPAGSPLPSSASPPAPPTSLTPDGAP 93
Query: 125 STGPTGARHFSNHNSSS 141
GPTG+ N+N+++
Sbjct: 94 VAGPTGSTPVDNNNAAT 110
>At1g80810 unknown protein
Length = 826
Score = 30.8 bits (68), Expect = 0.91
Identities = 13/26 (50%), Positives = 16/26 (61%)
Query: 153 DPLIGKKVWTRWPEDSHFYEAVITDY 178
+ L+GK+V WP D FYE VI Y
Sbjct: 556 EDLVGKRVNIWWPLDKTFYEGVIDSY 581
>At4g31880 unknown protein
Length = 873
Score = 30.4 bits (67), Expect = 1.2
Identities = 21/90 (23%), Positives = 34/90 (37%), Gaps = 14/90 (15%)
Query: 90 TVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPAEE 149
T S N+KP S L S + Q P N N+ + +
Sbjct: 557 TSSGDNEKPAVSSG-------KLASKSKKEAKQTVEESP-------NSNTKRKRSLGQGK 602
Query: 150 AQFDPLIGKKVWTRWPEDSHFYEAVITDYN 179
A + L+G ++ WP D +Y+ V+ Y+
Sbjct: 603 ASGESLVGSRIKVWWPMDQAYYKGVVESYD 632
>At5g10950 putative protein
Length = 395
Score = 30.0 bits (66), Expect = 1.6
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query: 132 RHFSNHNSSSNLNAPAEEAQF-DPLIGKKVWTRWPEDSHFYEAVITDY 178
R S+ SS + ++ ++ + L+G ++ WP DS FY+ V+ Y
Sbjct: 13 RLVSSSTRSSGKDKVSDARKYGEALVGSRIRVWWPMDSKFYKGVVDSY 60
>At1g36150 hypothetical protein
Length = 256
Score = 29.6 bits (65), Expect = 2.0
Identities = 23/69 (33%), Positives = 30/69 (43%), Gaps = 8/69 (11%)
Query: 79 SQPVHDILPSPTVSASNKK--------PKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTG 130
S PV + SPT S S+ K P SHS PP S P S PV +S +
Sbjct: 129 STPVSPPVESPTTSPSSAKSPAITPSSPAVSHSPPPVRHSSPPVSHSSPPVSHSSPPTSR 188
Query: 131 ARHFSNHNS 139
+ +H+S
Sbjct: 189 SSPAVSHSS 197
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 29.6 bits (65), Expect = 2.0
Identities = 25/93 (26%), Positives = 38/93 (39%), Gaps = 11/93 (11%)
Query: 73 PPGCTTSQ-----PVHDILPSPTVSASNKKPKTSHSVP------PSLGQSLPGLSSVKPV 121
PP T S+ PV PSP +S+ P + S P P L LP L+ P
Sbjct: 21 PPPETPSENSALPPVDSSPPSPPADSSSTPPLSEPSTPPPDSQLPPLPSILPPLTDSPPP 80
Query: 122 QCASTGPTGARHFSNHNSSSNLNAPAEEAQFDP 154
S+ P + +S+ +P E+++ P
Sbjct: 81 PSDSSPPVDSTPSPPPPTSNESPSPPEDSETPP 113
>At1g79480 hypothetical protein
Length = 356
Score = 29.3 bits (64), Expect = 2.7
Identities = 22/73 (30%), Positives = 30/73 (40%), Gaps = 2/73 (2%)
Query: 74 PGCTTSQPVHDILPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARH 133
P + PV +P+P S+SN P S S P S S+ P P + +
Sbjct: 118 PNSNPNPPV--TVPNPPESSSNPNPPDSSSNPNSNPNPPESSSNPNPPVTVPNPPESSSN 175
Query: 134 FSNHNSSSNLNAP 146
+ SSSN N P
Sbjct: 176 PNPPESSSNPNPP 188
>At5g53570 GTPase activator protein of Rab-like small GTPases-like
protein
Length = 550
Score = 28.9 bits (63), Expect = 3.5
Identities = 28/104 (26%), Positives = 41/104 (38%), Gaps = 26/104 (25%)
Query: 92 SASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSS---------- 141
S S+ +S S+P S SLP SS P + + +SN NSSS
Sbjct: 10 STSSGNSSSSSSLPSSSSSSLPSSSSSSPPS------SNSNSYSNSNSSSSSSSWIHLRS 63
Query: 142 -----NLNAPAEEAQFDPLIGKKVWTR----WPEDSHFYEAVIT 176
NL++P+ D K W+R W H + ++ T
Sbjct: 64 VLFVANLSSPSSVTSSDRR-RKSPWSRRKRKWALTPHQWRSLFT 106
>At3g60900 GPI-anchored protein (Fla10)
Length = 422
Score = 28.9 bits (63), Expect = 3.5
Identities = 19/69 (27%), Positives = 33/69 (47%), Gaps = 6/69 (8%)
Query: 80 QPVHDILPSPTVSASN-KKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHN 138
+PV P+P S S + P + + PP+ P + P S PT + + + N
Sbjct: 344 EPVSAPTPTPAKSPSPVEAPSPTAASPPA-----PPVDESSPEGAPSDSPTSSENSNAKN 398
Query: 139 SSSNLNAPA 147
++ ++NAPA
Sbjct: 399 AAFHVNAPA 407
>At1g60020 hypothetical protein
Length = 1194
Score = 28.9 bits (63), Expect = 3.5
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 5/68 (7%)
Query: 77 TTSQPVHDILPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSN 136
TT+ P + P+V + P H PPS Q P S + P +S T + FS+
Sbjct: 728 TTTVPSPPFVQLPSVPPPTRDP---HQPPPS--QPAPSPSPLSPPSMSSPVMTSSPQFSS 782
Query: 137 HNSSSNLN 144
+ S+ L+
Sbjct: 783 NRDSTTLH 790
>At1g49490 hypothetical protein
Length = 847
Score = 28.9 bits (63), Expect = 3.5
Identities = 17/62 (27%), Positives = 22/62 (35%)
Query: 70 IYQPPGCTTSQPVHDILPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPT 129
++ PP P + P S P SHS PP + P S P + P
Sbjct: 591 VHSPPPPPVFSPPPPVFSPPPPSPVYSPPPPSHSPPPPVYSPPPPTFSPPPTHNTNQPPM 650
Query: 130 GA 131
GA
Sbjct: 651 GA 652
>At1g70460 putative protein kinase
Length = 710
Score = 28.5 bits (62), Expect = 4.5
Identities = 19/61 (31%), Positives = 27/61 (44%), Gaps = 5/61 (8%)
Query: 73 PPGCTTSQ-----PVHDILPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTG 127
PP T+S P D P P+ A + P S+PP++ P +SS P S+
Sbjct: 18 PPPDTSSDGSAAPPPTDSAPPPSPPADSSPPPALPSLPPAVFSPPPTVSSPPPPPLDSSP 77
Query: 128 P 128
P
Sbjct: 78 P 78
>At5g55020 putative transcription factor MYB120 (MYB120)
Length = 523
Score = 28.1 bits (61), Expect = 5.9
Identities = 19/70 (27%), Positives = 30/70 (42%), Gaps = 1/70 (1%)
Query: 60 HRTRDWRETGIYQPPGCTTSQPVHDILPSPT-VSASNKKPKTSHSVPPSLGQSLPGLSSV 118
H+ + + +QP + P LPSPT +A + T H+ +L L +
Sbjct: 166 HQQQQQHQQMYFQPQSSQRNTPSSSPLPSPTPANAKSSSSFTFHTTTANLLHPLSPHTPN 225
Query: 119 KPVQCASTGP 128
P Q +ST P
Sbjct: 226 TPSQLSSTPP 235
>At4g32970 putative protein
Length = 743
Score = 28.1 bits (61), Expect = 5.9
Identities = 10/26 (38%), Positives = 16/26 (61%)
Query: 153 DPLIGKKVWTRWPEDSHFYEAVITDY 178
+ ++G +V WP D +YEAV+ Y
Sbjct: 276 EKIVGSRVKIWWPLDRAYYEAVVISY 301
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.335 0.147 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,757,805
Number of Sequences: 26719
Number of extensions: 291484
Number of successful extensions: 1596
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 1542
Number of HSP's gapped (non-prelim): 45
length of query: 296
length of database: 11,318,596
effective HSP length: 99
effective length of query: 197
effective length of database: 8,673,415
effective search space: 1708662755
effective search space used: 1708662755
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138087.2


