

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138087.10 + phase: 0
(242 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
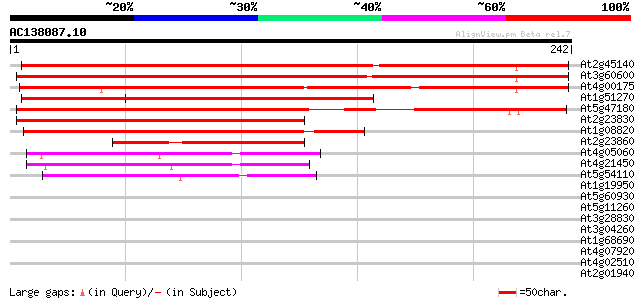
Score E
Sequences producing significant alignments: (bits) Value
At2g45140 putative VAMP-associated protein 329 7e-91
At3g60600 unknown protein 310 6e-85
At4g00175 putative proline-rich protein 280 4e-76
At1g51270 unknown protein 209 1e-54
At5g47180 VAMP (vesicle-associated membrane protein)-associated ... 184 5e-47
At2g23830 unknown protein 174 4e-44
At1g08820 unknown protein 149 1e-36
At2g23860 pseudogene 98 5e-21
At4g05060 unknown protein 68 4e-12
At4g21450 unknown protein 66 1e-11
At5g54110 membrane associated protein 62 3e-10
At1g19950 unknown protein 34 0.062
At5g60930 microtubule-associated motor - like 33 0.14
At5g11260 bZip transcription factor HY5 / AtbZip56 33 0.18
At3g28830 hypothetical protein 32 0.31
At3g04260 hypothetical protein 32 0.40
At1g68690 protein kinase, putative 31 0.68
At4g07920 putative protein 30 0.89
At4g02510 chloroplast protein import component Toc159-like 30 1.5
At2g01940 putative C2H2-type zinc finger protein 30 1.5
>At2g45140 putative VAMP-associated protein
Length = 239
Score = 329 bits (844), Expect = 7e-91
Identities = 160/237 (67%), Positives = 205/237 (85%), Gaps = 3/237 (1%)
Query: 6 DLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
+LL+I+P++L+FPFELKKQISCSL L NKTD+YVAFKVKTTNP+KYCVRPNTG+V PRS+
Sbjct: 4 ELLTIDPVDLQFPFELKKQISCSLYLGNKTDNYVAFKVKTTNPKKYCVRPNTGVVHPRSS 63
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVV 125
+V+VTMQAQKEAPAD+QCKDKFLLQ V + G +PKD++ EMF+KEAGH VEE KLRVV
Sbjct: 64 SEVLVTMQAQKEAPADLQCKDKFLLQCVVASPGATPKDVTHEMFSKEAGHRVEETKLRVV 123
Query: 126 YVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARALI 185
YV+PP+PPSPV EGSEEGSSPR S S+NGNA+ +F R SA+R +AQD S+EARAL+
Sbjct: 124 YVAPPRPPSPVREGSEEGSSPRASVSDNGNAS--DFTAAPRFSADRVDAQDNSSEARALV 181
Query: 186 SRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
++LTEEKN+A+Q +RL+QEL+ L+RE R++ GG+ F+ V+L+GL+GLI+GY+MK+
Sbjct: 182 TKLTEEKNSAVQLNNRLQQELDQLRRESKRSKSGGIPFMYVLLVGLIGLILGYIMKR 238
>At3g60600 unknown protein
Length = 256
Score = 310 bits (793), Expect = 6e-85
Identities = 153/239 (64%), Positives = 200/239 (83%), Gaps = 3/239 (1%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ +LL++EPL+L+FPFELKKQISCSL L+NKTD+ VAFKVKTTNP+KYCVRPNTG+VLPR
Sbjct: 19 NSELLTVEPLDLQFPFELKKQISCSLYLTNKTDNNVAFKVKTTNPKKYCVRPNTGVVLPR 78
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
STC+V+VTMQAQKEAP+DMQCKDKFLLQ V + GV+ K+++ EMF+KEAGH VEE KLR
Sbjct: 79 STCEVLVTMQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEMFSKEAGHRVEETKLR 138
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V YV+PP+PPSPV EGSEEGSSPR S S+NG +G EF+ + Q+ ++EARA
Sbjct: 139 VTYVAPPRPPSPVHEGSEEGSSPRASVSDNG--HGSEFSFERFIVDNKAGHQENTSEARA 196
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
LI++LTEEK +AIQ +RL++EL+ L+RE +++ GG+ F+ V+L+GL+GLI+GY+MK+
Sbjct: 197 LITKLTEEKQSAIQLNNRLQRELDQLRRESKKSQSGGIPFMYVLLVGLIGLILGYIMKR 255
>At4g00175 putative proline-rich protein
Length = 239
Score = 280 bits (717), Expect = 4e-76
Identities = 143/239 (59%), Positives = 188/239 (77%), Gaps = 6/239 (2%)
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSY-VAFKVKTTNPRKYCVRPNTGIVLPR 63
GDL++I P ELKFPFELKKQ SCS+QL+NKT + VAFKVKTTNPRKYCVRPNTG+VLP
Sbjct: 4 GDLVNIHPTELKFPFELKKQSSCSMQLTNKTTTQCVAFKVKTTNPRKYCVRPNTGVVLPG 63
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
+C+V VTMQAQKEAP DMQCKDKFL+Q+V +DG + K++ AEMFNKEAG V+E+ KLR
Sbjct: 64 DSCNVTVTMQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNKEAGRVIEDFKLR 123
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
VVY+ P PPSPVPEGSEEG+SP S ++ + + F V+R E E KS+EA +
Sbjct: 124 VVYI-PANPPSPVPEGSEEGNSPMASLNDIASQSASLFDDVSRTFEETSE---KSSEAWS 179
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
+IS+LTEEK +A QQ+ +LR ELE+L++E ++ + GG S ++++L+GLLG ++GYL+ +
Sbjct: 180 MISKLTEEKTSATQQSQKLRLELEMLRKETSKKQSGGHSLLLMLLVGLLGCVIGYLLNR 238
>At1g51270 unknown protein
Length = 289
Score = 209 bits (532), Expect = 1e-54
Identities = 96/152 (63%), Positives = 125/152 (82%)
Query: 6 DLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
+LL I+P++++FP EL K++SCSL L+NKT++YVAFK KTTN +KY VRPN G+VLPRS+
Sbjct: 127 ELLIIDPVDVQFPIELNKKVSCSLNLTNKTENYVAFKAKTTNAKKYYVRPNVGVVLPRSS 186
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVV 125
C+V+V MQA KEAPADMQC+DK L Q + KD+++EMF+KEAGH EE +L+V+
Sbjct: 187 CEVLVIMQALKEAPADMQCRDKLLFQCKVVEPETTAKDVTSEMFSKEAGHPAEETRLKVM 246
Query: 126 YVSPPQPPSPVPEGSEEGSSPRGSFSENGNAN 157
YV+PPQPPSPV EG+EEGSSPR S S+NGNA+
Sbjct: 247 YVTPPQPPSPVQEGTEEGSSPRASVSDNGNAS 278
Score = 139 bits (349), Expect = 2e-33
Identities = 63/107 (58%), Positives = 81/107 (74%)
Query: 51 YCVRPNTGIVLPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFN 110
YCV+P+ G+VLPRS+C+V+V MQA KEAPAD QCKDK L Q G K++++EMF+
Sbjct: 2 YCVKPSVGVVLPRSSCEVLVVMQALKEAPADRQCKDKLLFQCKVVEPGTMDKEVTSEMFS 61
Query: 111 KEAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNAN 157
KEAGH VEE +++YV+PPQP SPV EG E+GSSP S S+ GNA+
Sbjct: 62 KEAGHRVEETIFKIIYVAPPQPQSPVQEGLEDGSSPSASVSDKGNAS 108
>At5g47180 VAMP (vesicle-associated membrane protein)-associated
protein-like
Length = 220
Score = 184 bits (466), Expect = 5e-47
Identities = 101/239 (42%), Positives = 148/239 (61%), Gaps = 33/239 (13%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ L+SI+P ELKF FEL+KQ C L+++NKT++YVAFKVKTT+P+KY VRPNTG++ P
Sbjct: 6 ENQLISIQPDELKFLFELEKQSYCDLKVANKTENYVAFKVKTTSPKKYFVRPNTGVIQPW 65
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
+C + VT+QAQ+E P DMQCKDKFLLQS ++ + F K++G + ECKL+
Sbjct: 66 DSCIIRVTLQAQREYPPDMQCKDKFLLQSTIVPPHTDVDELPQDTFTKDSGKTLTECKLK 125
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V Y++P S SE+G NG +S+E +
Sbjct: 126 VSYITP---------------STTQRSSESGATNG----------------DGQSSETIS 154
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGN-RNRG-GVSFIIVILIGLLGLIMGYLMK 240
I RL EE++ A++QT +L+ ELE ++R N RN G G+S + ++GL+GLI+G+++K
Sbjct: 155 TIQRLKEERDAAVKQTQQLQHELETVRRRRNQRNSGNGLSLKLAAMVGLIGLIIGFILK 213
>At2g23830 unknown protein
Length = 149
Score = 174 bits (441), Expect = 4e-44
Identities = 82/124 (66%), Positives = 104/124 (83%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ +LL IEP+ L+FPFELKKQ+SCSL L+NKTD+ VAFKVKTTN YCVRPN G++LP+
Sbjct: 3 NNELLEIEPMYLQFPFELKKQMSCSLYLTNKTDNNVAFKVKTTNRNNYCVRPNYGLILPK 62
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
STC V+VTMQAQKE P+DMQ +KF++QSV + GV+ K+++ EMF+KE+GHVVEE KLR
Sbjct: 63 STCKVLVTMQAQKEVPSDMQSFEKFMIQSVLASPGVTAKEVTREMFSKESGHVVEETKLR 122
Query: 124 VVYV 127
V YV
Sbjct: 123 VTYV 126
>At1g08820 unknown protein
Length = 386
Score = 149 bits (376), Expect = 1e-36
Identities = 78/147 (53%), Positives = 99/147 (67%), Gaps = 4/147 (2%)
Query: 7 LLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTC 66
LL I+P L+F +LKKQ SC +QL+N T YVAFKVKTT+P+KYCVRPN G+V P+STC
Sbjct: 5 LLDIQPRTLQFAVDLKKQTSCVVQLTNTTHHYVAFKVKTTSPKKYCVRPNVGVVAPKSTC 64
Query: 67 DVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVY 126
+ V MQA KE P DM CKDKFL+QS + + +DI+A MF+K G +EE KLRV
Sbjct: 65 EFTVIMQAFKEPPPDMVCKDKFLIQSTAVSAETTDEDITASMFSKAEGKHIEENKLRVTL 124
Query: 127 VSPPQPPSPVPEGSEEGSSPRGSFSEN 153
V PPS PE S + +G+ E+
Sbjct: 125 V----PPSDSPELSPINTPKQGAVFED 147
>At2g23860 pseudogene
Length = 110
Score = 97.8 bits (242), Expect = 5e-21
Identities = 44/83 (53%), Positives = 61/83 (73%), Gaps = 5/83 (6%)
Query: 45 TTNPRKYCVRPNTGIVLPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDI 104
TTNP+ YCVRPN G++LP+STC + +K P+DMQ +KF++Q VK + V+ K++
Sbjct: 2 TTNPKNYCVRPNYGLILPKSTCKLK-----RKPGPSDMQSNEKFMIQRVKASPAVTAKEV 56
Query: 105 SAEMFNKEAGHVVEECKLRVVYV 127
+ E FNKE+GH+VEE KLRV YV
Sbjct: 57 TLETFNKESGHLVEETKLRVTYV 79
>At4g05060 unknown protein
Length = 287
Score = 68.2 bits (165), Expect = 4e-12
Identities = 43/132 (32%), Positives = 69/132 (51%), Gaps = 8/132 (6%)
Query: 8 LSIEP-LELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR--- 63
L ++P +L FP+E KQ+ ++++ N + S+VAFK +TT P+ +RP I+ P
Sbjct: 100 LKLDPSAKLYFPYEPGKQVRSAIKIKNTSKSHVAFKFQTTVPKSCFMRPAGAILAPGEEI 159
Query: 64 -STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKL 122
+T V E P + + KF + S+K P D E+F ++ HV EE +
Sbjct: 160 IATVFKFVEPPENNEKPMEQKSGVKFKIMSLKMK---VPTDYMPELFEEQKDHVSEEQVM 216
Query: 123 RVVYVSPPQPPS 134
RVV++ P P S
Sbjct: 217 RVVFLDPENPNS 228
>At4g21450 unknown protein
Length = 295
Score = 66.2 bits (160), Expect = 1e-11
Identities = 41/127 (32%), Positives = 67/127 (52%), Gaps = 8/127 (6%)
Query: 8 LSIEPLE-LKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTC 66
L ++P E L FP+E KQ+ ++++ N + S+VAFK +TT P+ +RP I+ P T
Sbjct: 108 LKLDPSEKLYFPYEPGKQVRSAIKIKNTSKSHVAFKFQTTAPKSCFMRPPGAILAPGETI 167
Query: 67 DV----MVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKL 122
V E P D + + KF + S+K P D E+F+++ V +E L
Sbjct: 168 IATVFKFVEPPENNEKPMDQRSRVKFKIMSLKVK---GPMDYVPELFDEQKDDVSKEQIL 224
Query: 123 RVVYVSP 129
RV+++ P
Sbjct: 225 RVIFLDP 231
>At5g54110 membrane associated protein
Length = 266
Score = 62.0 bits (149), Expect = 3e-10
Identities = 36/123 (29%), Positives = 65/123 (52%), Gaps = 8/123 (6%)
Query: 15 LKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCDVMVTM-- 72
L FP+E KQ+ +++L N + S+ AFK +TT P+ +RP G++ P + V
Sbjct: 85 LYFPYEPGKQVRSAIKLKNTSKSHTAFKFQTTAPKSCYMRPPGGVLAPGESVFATVFKFV 144
Query: 73 ---QAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYVSP 129
+ ++ P + + K KF + S+K GV + E+F+++ V E LRV+++
Sbjct: 145 EHPENNEKQPLNQKSKVKFKIMSLKVKPGV---EYVPELFDEQKDQVAVEQVLRVIFIDA 201
Query: 130 PQP 132
+P
Sbjct: 202 DRP 204
>At1g19950 unknown protein
Length = 315
Score = 34.3 bits (77), Expect = 0.062
Identities = 35/144 (24%), Positives = 57/144 (39%), Gaps = 21/144 (14%)
Query: 67 DVMVTMQAQKEAP-ADMQCKDKFLLQSVKTNDGVSPKDISA-------EMFNKEAGHVVE 118
D+ T QA P +Q K L+ T + +SPK +S+ + KEA V
Sbjct: 170 DLKATSQAASSNPQVRLQSKKPQLV----TKEPISPKPLSSPRKQQQLQTETKEAKASVS 225
Query: 119 ECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKS 178
+ KL + P PP P P S ++ R + + E A T + P ++
Sbjct: 226 QTKLTTLTPPGPPPPPPPPPPSPTTAAKRNADPAQPSPTEAEEASQTVAALPEPASE--- 282
Query: 179 AEARALISRLTEEKNNAIQQTSRL 202
I R + K +++T R+
Sbjct: 283 ------IQRASSSKETIMEETLRI 300
>At5g60930 microtubule-associated motor - like
Length = 647
Score = 33.1 bits (74), Expect = 0.14
Identities = 19/55 (34%), Positives = 28/55 (50%)
Query: 156 ANGPEFAQVTRGSAERPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLK 210
ANGP + + E + E R+ R TEE+ ++ +RLR+E ELLK
Sbjct: 110 ANGPGTQALMQAIEHEIEVTVRVHEVRSEYERQTEERARMAKEVARLREENELLK 164
>At5g11260 bZip transcription factor HY5 / AtbZip56
Length = 168
Score = 32.7 bits (73), Expect = 0.18
Identities = 21/75 (28%), Positives = 33/75 (44%)
Query: 136 VPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARALISRLTEEKNNA 195
VPE E S E+G+A G E Q T G ++R + + + + RL + +A
Sbjct: 43 VPEFGGEAVGKETSGRESGSATGQERTQATVGESQRKRGRTPAEKENKRLKRLLRNRVSA 102
Query: 196 IQQTSRLRQELELLK 210
Q R + L L+
Sbjct: 103 QQARERKKAYLSELE 117
>At3g28830 hypothetical protein
Length = 536
Score = 32.0 bits (71), Expect = 0.31
Identities = 22/79 (27%), Positives = 35/79 (43%), Gaps = 1/79 (1%)
Query: 128 SPPQPPSPVPEGSEEG-SSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARALIS 186
SP P P GS G SS +GS S G+A+ A ++ + A + E+ A+
Sbjct: 277 SPSGSPKASPSGSVSGKSSSKGSASAQGSASAQGSASAQGSASAQGSASAQRRESGAMAM 336
Query: 187 RLTEEKNNAIQQTSRLRQE 205
+ E + Q+ S+ E
Sbjct: 337 SKSRETKTSSQRQSKSSSE 355
>At3g04260 hypothetical protein
Length = 913
Score = 31.6 bits (70), Expect = 0.40
Identities = 29/110 (26%), Positives = 53/110 (47%), Gaps = 13/110 (11%)
Query: 122 LRVVYVSPPQPPS---PVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKS 178
+ +++++PP P + P+P + SS R S S A + R + +
Sbjct: 1 MSLLFLNPPFPSNSIHPIPRRAAGISSIRCSIS----------APEKKPRRRRKQKRGDG 50
Query: 179 AEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILI 228
AE +S + E +A++++ RL EL++R NR+ GVS +I +I
Sbjct: 51 AENDDSLSFGSGEAVSALERSLRLTFMDELMERARNRDTSGVSEVIYDMI 100
>At1g68690 protein kinase, putative
Length = 708
Score = 30.8 bits (68), Expect = 0.68
Identities = 31/124 (25%), Positives = 47/124 (37%), Gaps = 10/124 (8%)
Query: 128 SPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDK----SAEARA 183
SPP PPS P S SP S+ + + P + T+ R S+ R+
Sbjct: 183 SPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPRRSPNSPPPTFSSPPRS 242
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNRG-----GVSFIIVILI-GLLGLIMGY 237
L NN Q LR L+ N G G+S + +++ L G+ +
Sbjct: 243 PPEILVPGSNNPSQNNPTLRPPLDAPNSTNNSGIGTGAVVGISVAVALVVFTLFGIFVWC 302
Query: 238 LMKK 241
L K+
Sbjct: 303 LRKR 306
>At4g07920 putative protein
Length = 314
Score = 30.4 bits (67), Expect = 0.89
Identities = 33/137 (24%), Positives = 49/137 (35%), Gaps = 21/137 (15%)
Query: 51 YCVRPNTGIVLPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFN 110
YC+ + P C V+ +E + C D L VKTN S +
Sbjct: 90 YCLGFAFPHMCPNFVCHVLALFMRAQEINKEFGCHDLLQLCQVKTNAR------SDQGTK 143
Query: 111 KEAGHVVEECKLRVVYVSPPQPPSP--------VPEGSEEGSSPRGSFSENGNANGPEFA 162
+ G+ V+ +P PSP + +GS S R S S + +
Sbjct: 144 NQRGYPVKN-------GTPAVNPSPTWSDRLGHIQQGSHSSCSRRSSSSSSNCPSSSAAR 196
Query: 163 QVTRGSAERPEAQDKSA 179
RG A R E + +SA
Sbjct: 197 SDRRGPAGRTEPRGRSA 213
>At4g02510 chloroplast protein import component Toc159-like
Length = 1503
Score = 29.6 bits (65), Expect = 1.5
Identities = 20/95 (21%), Positives = 41/95 (43%), Gaps = 5/95 (5%)
Query: 107 EMFNKEAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTR 166
E F E+ + + KL V S +P +E GS P+ NG G +A+V +
Sbjct: 297 EKFTSESDSIADSSKLESVDTSAVEPEVVA---AESGSEPKDVEKANGLEKGMTYAEVIK 353
Query: 167 GSAERPEAQDKSAEARALISRLTEEKNNAIQQTSR 201
A A + + E +++ + ++ ++ ++
Sbjct: 354 --AASAVADNGTKEEESVLGGIVDDAEEGVKLNNK 386
>At2g01940 putative C2H2-type zinc finger protein
Length = 439
Score = 29.6 bits (65), Expect = 1.5
Identities = 30/102 (29%), Positives = 38/102 (36%), Gaps = 16/102 (15%)
Query: 118 EECKLRVVYVSPPQPPS---PVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEA 174
+ C R V+ PP+PP VP S +S + S N G + RP
Sbjct: 191 DNCSARRVHREPPRPPQTAVTVPACSSRTASTVSTPSSETNYGGTVAVTTPQPLEGRPIH 250
Query: 175 QDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRN 216
Q R S LT NN L EL+LL N+N
Sbjct: 251 Q------RISSSILTNSSNN-------LNLELQLLPLSSNQN 279
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,579,678
Number of Sequences: 26719
Number of extensions: 244736
Number of successful extensions: 1140
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 1067
Number of HSP's gapped (non-prelim): 85
length of query: 242
length of database: 11,318,596
effective HSP length: 96
effective length of query: 146
effective length of database: 8,753,572
effective search space: 1278021512
effective search space used: 1278021512
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC138087.10


