

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138063.3 - phase: 0 /pseudo
(111 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
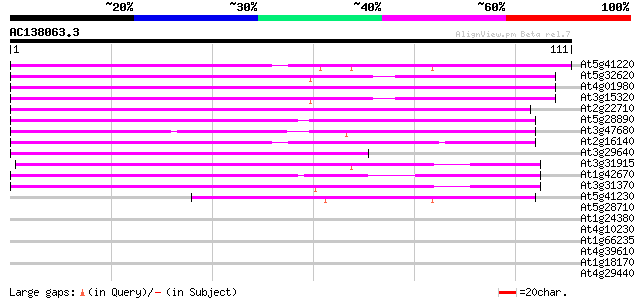
Score E
Sequences producing significant alignments: (bits) Value
At5g41220 glutathione transferase-like 70 2e-13
At5g32620 putative protein 67 1e-12
At4g01980 hypothetical protein 66 2e-12
At3g15320 hypothetical protein 66 2e-12
At2g22710 hypothetical protein 65 4e-12
At5g28890 putative protein 64 9e-12
At3g47680 unknown protein 62 4e-11
At2g16140 pseudogene; similar to MURA transposase of maize Muta... 62 5e-11
At3g29640 hypothetical protein 62 6e-11
At3g31915 hypothetical protein 55 4e-09
At1g42670 unknown protein 51 1e-07
At3g31370 hypothetical protein 49 4e-07
At5g41230 putative protein 42 4e-05
At5g28710 putative protein 38 0.001
At1g24380 hypothetical protein 36 0.003
At4g10230 hypothetical protein 35 0.005
At1g66235 putative protein 35 0.008
At4g39610 unknown protein 33 0.030
At1g18170 unknown protein 32 0.040
At4g29440 putative protein 28 0.57
>At5g41220 glutathione transferase-like
Length = 590
Score = 70.1 bits (170), Expect = 2e-13
Identities = 44/124 (35%), Positives = 62/124 (49%), Gaps = 16/124 (12%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVGCY + + SG SE D A +Y + + FTLE+AW+ LR KW C L P
Sbjct: 342 FVGCYDQALNQRSSGQSEDDVFQVAYQVYTNNYKSNFTLEHAWRELRHSKKW---CSLYP 398
Query: 61 --------SAKRTK-NSASGAYLASSNPPTP----TSEYNPPQLPIGQKAAKKKGKEKHV 107
S+KRTK N+ Y +SSNP + E LP+G K++K+K K+
Sbjct: 399 FENSKGGGSSKRTKLNNGDRVYSSSSNPESVPIALDEEEQVMDLPLGVKSSKQKEKKVAT 458
Query: 108 VFQV 111
+ +
Sbjct: 459 IITI 462
>At5g32620 putative protein
Length = 301
Score = 67.0 bits (162), Expect = 1e-12
Identities = 40/110 (36%), Positives = 57/110 (51%), Gaps = 6/110 (5%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVL-- 58
FV CYK K SG SE D + A +IY+ D ++ FTL++AW+ LR + KW
Sbjct: 126 FVACYKTTTTHKTSGQSEDDVMKLAYEIYFNDTKKNFTLDHAWRELRYDQKWCEATSRKG 185
Query: 59 EPSAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKEKHVV 108
+ +AKR K A S+ P + + P G KAAK KG++ +V
Sbjct: 186 DENAKRRKCGDGNA----SSHPIHVEDDSIMSRPPGVKAAKAKGRKSAIV 231
>At4g01980 hypothetical protein
Length = 302
Score = 66.2 bits (160), Expect = 2e-12
Identities = 35/108 (32%), Positives = 57/108 (52%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVGCY K SG +E+D + A +++ D+++KF+L++AW+LLR + KWL +
Sbjct: 134 FVGCYATTTNQKASGQTENDVMLFANELFENDMKKKFSLDHAWRLLRHDQKWLISNAPKG 193
Query: 61 SAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKEKHVV 108
+ A+S+ P + + + P G KAAK K K+ V
Sbjct: 194 KGIAKRRKVRVGTQAASSQPIDLEDDDVMRRPPGVKAAKAKAKKTPTV 241
>At3g15320 hypothetical protein
Length = 287
Score = 66.2 bits (160), Expect = 2e-12
Identities = 39/110 (35%), Positives = 56/110 (50%), Gaps = 6/110 (5%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVL-- 58
FVGCYK K SG SE D + A +IY+ D ++ FTL++AW+ LR + KW
Sbjct: 112 FVGCYKTTTTHKTSGQSEDDVMKLAYEIYFNDTKKNFTLDHAWRELRYDQKWCEATSRKG 171
Query: 59 EPSAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKEKHVV 108
+ +AKR K A S+ P + + P G KAAK K ++ +
Sbjct: 172 DKNAKRKKCGDGNA----SSQPIHVEDDSVMSRPPGVKAAKAKARKSATI 217
>At2g22710 hypothetical protein
Length = 300
Score = 65.5 bits (158), Expect = 4e-12
Identities = 34/103 (33%), Positives = 55/103 (53%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVGCY K SG +E+D + A +++ D+++KF+L++AW+LLR + KWL +
Sbjct: 132 FVGCYATTTNQKASGQTENDVMLFANELFKNDMKKKFSLDHAWRLLRHDQKWLISNAPKG 191
Query: 61 SAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGK 103
+ A+S+ P + + + P G KAAK K K
Sbjct: 192 KGIAKRRKVRVGTQAASSQPIDLEDDDVMRRPPGVKAAKAKAK 234
>At5g28890 putative protein
Length = 232
Score = 64.3 bits (155), Expect = 9e-12
Identities = 34/104 (32%), Positives = 54/104 (51%), Gaps = 2/104 (1%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVGCY K SG +E+D + A +++ D+++KF+L++AW+L+R E KW+ P
Sbjct: 116 FVGCYATETNQKASGQTENDVMLFANELFENDMKKKFSLDHAWRLVRHEQKWIISNT--P 173
Query: 61 SAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKE 104
KR A S+ P +Y+ P+ K K K K+
Sbjct: 174 KEKRMSKRRKVGSQAKSSQPINLEDYDVMARPLRVKVPKAKTKK 217
>At3g47680 unknown protein
Length = 302
Score = 62.4 bits (150), Expect = 4e-11
Identities = 40/105 (38%), Positives = 56/105 (53%), Gaps = 6/105 (5%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVG Y+ K SG ++ D ++ A +IY + KFTLE+AW++LR E KWL+ P
Sbjct: 119 FVGSYEAATKQKSSGQNDDDVVALAHEIYNSE-HGKFTLEHAWRVLRFEQKWLSA----P 173
Query: 61 SAKRT-KNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKE 104
S K T + + AS++ P PIG KAAK K K+
Sbjct: 174 STKATVMSKRRKSDKASTSQPQTHEAEEAMSRPIGVKAAKAKAKK 218
>At2g16140 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 311
Score = 62.0 bits (149), Expect = 5e-11
Identities = 38/104 (36%), Positives = 54/104 (51%), Gaps = 4/104 (3%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVG Y+ K SG +++D IS A +I+ D KF LE+AW++LR + KW C
Sbjct: 129 FVGSYEAASREKSSGQNDNDVISLAHEIFNNDYGYKFPLEHAWRVLRHDQKW---CSQAS 185
Query: 61 SAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKE 104
+ + A ++S PP+ E PIG KAAK K K+
Sbjct: 186 VMSKRRKCDKAAQPSTSQPPSHGVE-EAMSRPIGVKAAKAKAKK 228
>At3g29640 hypothetical protein
Length = 171
Score = 61.6 bits (148), Expect = 6e-11
Identities = 27/71 (38%), Positives = 38/71 (53%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVGCYK K SG SE D + A +IYY D ++KF LE+ W+ LR + KW ++
Sbjct: 100 FVGCYKTATTHKTSGQSEDDVMKLAYEIYYNDTKKKFNLEHTWRKLRYDQKWCEAATIKG 159
Query: 61 SAKRTKNSASG 71
+ + G
Sbjct: 160 NKNAKRKCVDG 170
>At3g31915 hypothetical protein
Length = 239
Score = 55.5 bits (132), Expect = 4e-09
Identities = 35/105 (33%), Positives = 52/105 (49%), Gaps = 8/105 (7%)
Query: 2 VGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEPS 61
+G Y K SG +++D ++ + +Y D + KFTLE+AW+ +R + KW + L
Sbjct: 64 MGSYVAATKEKTSGQNDNDVLNFSHQSFYTDYKPKFTLEHAWREVRVDQKWCSSNALNVP 123
Query: 62 AKRTK-NSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKEK 105
AKR K N + G ++ T PIG KA K K K K
Sbjct: 124 AKRRKQNESVGDHVTIDLEGDQTR-------PIGVKATKGKAKAK 161
>At1g42670 unknown protein
Length = 579
Score = 50.8 bits (120), Expect = 1e-07
Identities = 36/105 (34%), Positives = 47/105 (44%), Gaps = 10/105 (9%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
FVG Y + SG ++ D +S A I+ +D KFT E+AW+ R + KW+A
Sbjct: 409 FVGSYLAASKQQTSGQNDDDVVSLAHQIFSKDYGCKFTCEHAWRERRYDQKWIAQST-HG 467
Query: 61 SAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKEK 105
AKR K A P + PIG KAAK K K
Sbjct: 468 KAKRRKCEADS---------EPVGVEDKEARPIGVKAAKAAAKAK 503
>At3g31370 hypothetical protein
Length = 272
Score = 48.9 bits (115), Expect = 4e-07
Identities = 32/106 (30%), Positives = 51/106 (47%), Gaps = 8/106 (7%)
Query: 1 FVGCYKDVVALKKSGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLE- 59
FVG Y K SG +++D ++ A ++ D + K TLE+ W+ + + KW + L
Sbjct: 92 FVGSYVATTKEKTSGENDNDVMNFAHHSFHTDYKTKLTLEHVWREVMFDQKWCSSNALNV 151
Query: 60 PSAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKGKEK 105
P+ +R +N + G ++ T PI KAAK K K K
Sbjct: 152 PAKQRKQNESVGDHVTVDLEGYQTR-------PIDVKAAKAKAKGK 190
>At5g41230 putative protein
Length = 276
Score = 42.4 bits (98), Expect = 4e-05
Identities = 27/77 (35%), Positives = 39/77 (50%), Gaps = 9/77 (11%)
Query: 37 FTLENAWKLLRDEPKWLAGCVLEPS-----AKRTKNSASGAYLASSNPPTP----TSEYN 87
F LE+AW+ LR KW + E S +KRTK + G Y +S NP + E
Sbjct: 4 FKLEHAWRELRHNKKWCSTYTSENSKGGGSSKRTKLNGGGVYSSSCNPESVPIALDGEEQ 63
Query: 88 PPQLPIGQKAAKKKGKE 104
P+G K++K+K K+
Sbjct: 64 VMDRPLGVKSSKQKEKK 80
>At5g28710 putative protein
Length = 264
Score = 37.7 bits (86), Expect = 0.001
Identities = 22/56 (39%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query: 14 SGASESDNISAAKDIYYQDVREKFTLENAWKLLRDEPKWLAGCVLEPSAKRTKNSA 69
SG ++ +S A I+ +D KFT E+AW+ LR + KW+A AKR K A
Sbjct: 114 SGQNDDFVVSLAHQIFSKDYGCKFTCEHAWRELRYDQKWIAQST-HGKAKRRKCEA 168
>At1g24380 hypothetical protein
Length = 273
Score = 36.2 bits (82), Expect = 0.003
Identities = 28/117 (23%), Positives = 51/117 (42%), Gaps = 18/117 (15%)
Query: 3 GCYKDVVALKKSGASESDNISAAKDI--YYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
GC + SGASE D ++ AK + Y+ + F ++ W +L+ K+ + P
Sbjct: 13 GCVNQIENKNPSGASEEDILNQAKMLLTQYEKYKRGFKFDHVWPILKGIEKFANDNMKTP 72
Query: 61 SA----KRTKNSASGAYLASSNPPTP------------TSEYNPPQLPIGQKAAKKK 101
+A R S+S + + + P+P + ++ P+G K AK+K
Sbjct: 73 AAFQGEGRDVTSSSSFSINTESSPSPGMNSIDLNMDSEDANFSLSSRPMGLKKAKRK 129
>At4g10230 hypothetical protein
Length = 273
Score = 35.4 bits (80), Expect = 0.005
Identities = 28/117 (23%), Positives = 50/117 (41%), Gaps = 18/117 (15%)
Query: 3 GCYKDVVALKKSGASESDNISAAKDI--YYQDVREKFTLENAWKLLRDEPKWLAGCVLEP 60
GC + SGASE D ++ AK + Y+ + F ++ W +L+ K+ + P
Sbjct: 13 GCVNQIENKNPSGASEEDILNQAKMLLTQYEKYKRGFKFDHVWPILKGIEKFANDNMKTP 72
Query: 61 SA----KRTKNSASGAYLASSNPPTP------------TSEYNPPQLPIGQKAAKKK 101
A R S+S + + + P+P + ++ P+G K AK+K
Sbjct: 73 PAFQGEGRDVTSSSSFSINTESSPSPGMNSIDLNMDSEDANFSLSSRPMGLKKAKRK 129
>At1g66235 putative protein
Length = 265
Score = 34.7 bits (78), Expect = 0.008
Identities = 32/120 (26%), Positives = 52/120 (42%), Gaps = 15/120 (12%)
Query: 4 CYKDVVALKKSGASESDNISAAKDIYYQDVREK--FTLENAWKLLRDEPKWLAG------ 55
C K + SGAS D + AK++ QD K + ++ W ++R+ K+ G
Sbjct: 59 CIKLCENRRSSGASSDDIFNQAKEMLMQDKHFKSGWKFDHVWNIIRNFEKFKDGATQAKK 118
Query: 56 ----CVLE-PSAKRTKNSASGAYLASSN--PPTPTSEYNPPQLPIGQKAAKKKGKEKHVV 108
C LE P+ ++SG++ S N +P Q PI K +K K K ++
Sbjct: 119 VLNLCGLENPTLDSVSQASSGSFSFSLNLDDEDDIIGRSPYQRPIRVKKSKLKRKNDQIL 178
>At4g39610 unknown protein
Length = 264
Score = 32.7 bits (73), Expect = 0.030
Identities = 20/49 (40%), Positives = 30/49 (60%), Gaps = 2/49 (4%)
Query: 65 TKNSASGAYLASSNP-PTPTSEYNPP-QLPIGQKAAKKKGKEKHVVFQV 111
T +S+S + + S+P P+PTS PP Q P+ Q + KK K + VF+V
Sbjct: 10 TSSSSSSSSIPPSSPTPSPTSTTTPPRQHPLLQPPSSKKKKNRTNVFRV 58
>At1g18170 unknown protein
Length = 247
Score = 32.3 bits (72), Expect = 0.040
Identities = 15/35 (42%), Positives = 21/35 (59%)
Query: 73 YLASSNPPTPTSEYNPPQLPIGQKAAKKKGKEKHV 107
Y +SSNPP P S PP P Q A ++ ++K+V
Sbjct: 28 YASSSNPPEPESSSPPPPPPPPQPLASQQKRKKNV 62
>At4g29440 putative protein
Length = 1071
Score = 28.5 bits (62), Expect = 0.57
Identities = 13/44 (29%), Positives = 28/44 (63%)
Query: 59 EPSAKRTKNSASGAYLASSNPPTPTSEYNPPQLPIGQKAAKKKG 102
+P+AK+ +++S ++L ++ +P + P LP + AAK++G
Sbjct: 945 KPAAKQRGSASSLSFLPKTDKASPDQDSPPKLLPKEKPAAKQQG 988
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.128 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,768,415
Number of Sequences: 26719
Number of extensions: 112227
Number of successful extensions: 506
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 457
Number of HSP's gapped (non-prelim): 56
length of query: 111
length of database: 11,318,596
effective HSP length: 87
effective length of query: 24
effective length of database: 8,994,043
effective search space: 215857032
effective search space used: 215857032
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC138063.3


