

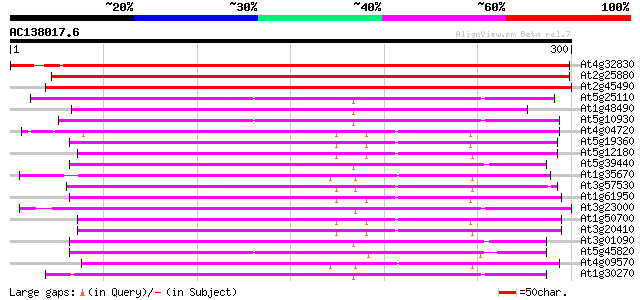
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138017.6 - phase: 0
(300 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g32830 putative serine/threonine protein kinase 541 e-154
At2g25880 putative protein kinase 524 e-149
At2g45490 putative protein kinase 373 e-104
At5g25110 serine/threonine protein kinase-like protein 176 1e-44
At1g48490 IRE - like protein kinase 176 1e-44
At5g10930 serine/threonine protein kinase -like protein 176 1e-44
At4g04720 putative calcium dependent protein kinase 171 3e-43
At5g19360 calcium-dependent protein kinase - like 170 8e-43
At5g12180 calcium-dependent protein kinase 170 1e-42
At5g39440 serine/threonine-specific protein kinase -like protein 169 2e-42
At1g35670 calcium-dependent protein kinase 168 3e-42
At3g57530 calcium-dependent protein kinase 168 4e-42
At1g61950 hypothetical protein 168 4e-42
At3g23000 SNF1 related protein kinase (ATSRPK1) 167 5e-42
At1g50700 hypothetical protein 167 5e-42
At3g20410 calmodulin-domain protein kinase CDPK isoform 9 167 8e-42
At3g01090 putative SNF1-related protein kinase 167 8e-42
At5g45820 CBL-interacting protein kinase 20 (CIPK20) 165 2e-41
At4g09570 calmodulin-domain protein kinase CDPK isoform 4 (CPK4) 165 2e-41
At1g30270 serine/threonine kinase, putative 165 2e-41
>At4g32830 putative serine/threonine protein kinase
Length = 294
Score = 541 bits (1393), Expect = e-154
Identities = 264/299 (88%), Positives = 282/299 (94%), Gaps = 6/299 (2%)
Query: 1 MAIATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSN 60
MAI TETQ Q + +S+ S +A Q+RW L+DFDIGKPLGRGKFGHVYLAREK SN
Sbjct: 1 MAIPTETQHQEKE-----ASDASAAAA-QKRWTLSDFDIGKPLGRGKFGHVYLAREKRSN 54
Query: 61 HIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKG 120
H+VALKVLFKSQLQQSQVEHQLRREVEIQSHLRHP+ILRLYGYFYDQKRVYLILEYA +G
Sbjct: 55 HVVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPNILRLYGYFYDQKRVYLILEYAARG 114
Query: 121 ELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFG 180
ELYK+LQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFG
Sbjct: 115 ELYKDLQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFG 174
Query: 181 WSVHTFNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDT 240
WSVHTFNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLG+LCYEFLYGVPPFEA EHSDT
Sbjct: 175 WSVHTFNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGILCYEFLYGVPPFEAMEHSDT 234
Query: 241 YRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAEPSGIYR 299
YRRI+QVDLKFPPKPI+S++AKDLISQMLVK+SS+RLPLHKLLEHPWIVQNA+PSGIYR
Sbjct: 235 YRRIVQVDLKFPPKPIISASAKDLISQMLVKESSQRLPLHKLLEHPWIVQNADPSGIYR 293
>At2g25880 putative protein kinase
Length = 282
Score = 524 bits (1349), Expect = e-149
Identities = 250/277 (90%), Positives = 268/277 (96%)
Query: 23 SGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQL 82
+ S Q+RW +DFDIGKPLGRGKFGHVYLAREK S+HIVALKVLFK+QLQQSQVEHQL
Sbjct: 5 AASEAAQKRWTTSDFDIGKPLGRGKFGHVYLAREKRSDHIVALKVLFKAQLQQSQVEHQL 64
Query: 83 RREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVA 142
RREVEIQSHLRHP+ILRLYGYFYDQKRVYLILEYA +GELYKELQKCKYFSERRAATYVA
Sbjct: 65 RREVEIQSHLRHPNILRLYGYFYDQKRVYLILEYAVRGELYKELQKCKYFSERRAATYVA 124
Query: 143 SLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPE 202
SLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPE
Sbjct: 125 SLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPE 184
Query: 203 MVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAK 262
MVESVEHDASVDIWSLG+LCYEFLYGVPPFEA+EHS+TY+RI+QVDLKFPPKPIVSS+AK
Sbjct: 185 MVESVEHDASVDIWSLGILCYEFLYGVPPFEAREHSETYKRIVQVDLKFPPKPIVSSSAK 244
Query: 263 DLISQMLVKDSSERLPLHKLLEHPWIVQNAEPSGIYR 299
DLISQMLVK+S++RL LHKLLEHPWIVQNA+PSG+YR
Sbjct: 245 DLISQMLVKESTQRLALHKLLEHPWIVQNADPSGLYR 281
>At2g45490 putative protein kinase
Length = 288
Score = 373 bits (957), Expect = e-104
Identities = 172/281 (61%), Positives = 230/281 (81%)
Query: 20 SEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVE 79
S S + +++W L DF+IG+PLG+GKFG VYLARE S +IVALKV+FK Q+++ ++
Sbjct: 5 STESDAGNTEKQWSLADFEIGRPLGKGKFGRVYLAREAKSKYIVALKVIFKEQIEKYKIH 64
Query: 80 HQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAAT 139
HQLRRE+EIQ+ LRHP+ILRL+G+F+D +R++LILEYA GELY L++ + +E++AAT
Sbjct: 65 HQLRREMEIQTSLRHPNILRLFGWFHDNERIFLILEYAHGGELYGVLKQNGHLTEQQAAT 124
Query: 140 YVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYL 199
Y+ASL++AL YCHGK VIHRDIKPENLL+ +G LKIADFGWSV + N+R+TMCGTLDYL
Sbjct: 125 YIASLSQALAYCHGKCVIHRDIKPENLLLDHEGRLKIADFGWSVQSSNKRKTMCGTLDYL 184
Query: 200 PPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSS 259
PEMVE+ +HD +VD W+LG+LCYEFLYG PPFEA+ DT++RI+++DL FP P VS
Sbjct: 185 APEMVENRDHDYAVDNWTLGILCYEFLYGNPPFEAESQKDTFKRILKIDLSFPLTPNVSE 244
Query: 260 AAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAEPSGIYRS 300
AK+LISQ+LVKD S+RL + K+++HPWIV+NA+P G+ S
Sbjct: 245 EAKNLISQLLVKDPSKRLSIEKIMQHPWIVKNADPKGVCAS 285
>At5g25110 serine/threonine protein kinase-like protein
Length = 488
Score = 176 bits (447), Expect = 1e-44
Identities = 96/286 (33%), Positives = 159/286 (55%), Gaps = 9/286 (3%)
Query: 12 HQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKS 71
H S+ + Q R + +++G+ LG+G FG VY +E T+ VA+K++ K
Sbjct: 18 HPDSRYQSAPTMEEEQQQLRVLFAKYEMGRLLGKGTFGKVYYGKEITTGESVAIKIINKD 77
Query: 72 QLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKY 131
Q+++ + Q++RE+ I +RHP+I+ L + +++ I+EY GEL+ ++ K K
Sbjct: 78 QVKREGMMEQIKREISIMRLVRHPNIVELKEVMATKTKIFFIMEYVKGGELFSKIVKGK- 136
Query: 132 FSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWS-----VHTF 186
E A Y L A+ +CH + V HRD+KPENLL+ G+LK++DFG S +
Sbjct: 137 LKEDSARKYFQQLISAVDFCHSRGVSHRDLKPENLLVDENGDLKVSDFGLSALPEQILQD 196
Query: 187 NRRRTMCGTLDYLPPEMVESVEHD-ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRII 245
T CGT Y+ PE++ +D A DIWS G++ Y L G PF+ + YR+I
Sbjct: 197 GLLHTQCGTPAYVAPEVLRKKGYDGAKGDIWSCGIILYVLLAGFLPFQDENLMKMYRKIF 256
Query: 246 QVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQN 291
+ + ++P P S +K LIS++LV D ++R+ + ++ PW +N
Sbjct: 257 KSEFEYP--PWFSPESKRLISKLLVVDPNKRISIPAIMRTPWFRKN 300
>At1g48490 IRE - like protein kinase
Length = 1235
Score = 176 bits (447), Expect = 1e-44
Identities = 93/273 (34%), Positives = 149/273 (54%), Gaps = 29/273 (10%)
Query: 34 LNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR 93
++DF++ K + RG FGHV LAR+ T+ + A+KVL K+ + + + E +I + R
Sbjct: 825 IDDFEVMKSISRGAFGHVILARKNTTGDLFAIKVLRKADMIRKNAVESILAERDILINAR 884
Query: 94 HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHG 153
+P ++R + F + +YL++EY G+ Y L+K E A Y+A + AL Y H
Sbjct: 885 NPFVVRFFYSFTCSENLYLVMEYLNGGDFYSMLRKIGCLDEANARVYIAEVVLALEYLHS 944
Query: 154 KHVIHRDIKPENLLIGAQGELKIADFGWS----------------------------VHT 185
+ V+HRD+KP+NLLI G +K+ DFG S + T
Sbjct: 945 EGVVHRDLKPDNLLIAHDGHVKLTDFGLSKVGLINNTDDLSGPVSSATSLLVEEKPKLPT 1004
Query: 186 FNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRII 245
+ +R+ GT DYL PE++ H A+ D WS+G++ YEFL G+PPF A + I+
Sbjct: 1005 LDHKRSAVGTPDYLAPEILLGTGHGATADWWSVGIILYEFLVGIPPFNADHPQQIFDNIL 1064
Query: 246 QVDLKFPPKP-IVSSAAKDLISQMLVKDSSERL 277
++++PP P +S A+DLI ++L +D +RL
Sbjct: 1065 NRNIQWPPVPEDMSHEARDLIDRLLTEDPHQRL 1097
>At5g10930 serine/threonine protein kinase -like protein
Length = 445
Score = 176 bits (446), Expect = 1e-44
Identities = 97/275 (35%), Positives = 157/275 (56%), Gaps = 10/275 (3%)
Query: 27 KDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQ-LQQSQVEHQLRRE 85
+++RR + +++G+ LG+G F VY +E VA+KV+ K Q +++ + Q++RE
Sbjct: 2 EEERRVLFGKYEMGRLLGKGTFAKVYYGKEIIGGECVAIKVINKDQVMKRPGMMEQIKRE 61
Query: 86 VEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLA 145
+ I +RHP+I+ L + +++ ++E+ GEL+ ++ K K E A Y L
Sbjct: 62 ISIMKLVRHPNIVELKEVMATKTKIFFVMEFVKGGELFCKISKGK-LHEDAARRYFQQLI 120
Query: 146 RALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWS-----VHTFNRRRTMCGTLDYLP 200
A+ YCH + V HRD+KPENLL+ G+LKI+DFG S + T CGT Y+
Sbjct: 121 SAVDYCHSRGVSHRDLKPENLLLDENGDLKISDFGLSALPEQILQDGLLHTQCGTPAYVA 180
Query: 201 PEMVESVEHD-ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSS 259
PE+++ +D A DIWS GV+ Y L G PF+ + + YR+I + D +FP P S
Sbjct: 181 PEVLKKKGYDGAKADIWSCGVVLYVLLAGCLPFQDENLMNMYRKIFRADFEFP--PWFSP 238
Query: 260 AAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAEP 294
A+ LIS++LV D R+ + ++ PW+ +N P
Sbjct: 239 EARRLISKLLVVDPDRRISIPAIMRTPWLRKNFTP 273
>At4g04720 putative calcium dependent protein kinase
Length = 531
Score = 171 bits (434), Expect = 3e-43
Identities = 105/303 (34%), Positives = 165/303 (53%), Gaps = 18/303 (5%)
Query: 7 TQPQPHQQHHTASSEVSGSAKDQRRWILNDFD-------IGKPLGRGKFGHVYLAREKTS 59
TQP HQQ T SS S +D + F+ +GK LGRG+FG Y+ +E +
Sbjct: 45 TQPI-HQQISTPSSNPV-SVRDPDTILGKPFEDIRKFYSLGKELGRGQFGITYMCKEIGT 102
Query: 60 NHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR-HPHILRLYGYFYDQKRVYLILEYAP 118
+ A K + K +L Q + ++RE++I +L P+I+ + G + D++ ++L++E
Sbjct: 103 GNTYACKSILKRKLISKQDKEDVKREIQIMQYLSGQPNIVEIKGAYEDRQSIHLVMELCA 162
Query: 119 KGELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGE---LK 175
GEL+ + ++SER AA + S+ + CH V+HRD+KPEN L+ ++ E LK
Sbjct: 163 GGELFDRIIAQGHYSERAAAGIIRSIVNVVQICHFMGVVHRDLKPENFLLSSKEENAMLK 222
Query: 176 IADFGWSVHTFNRR--RTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFE 233
DFG SV + R + G+ Y+ PE++ + +DIWS GV+ Y L GVPPF
Sbjct: 223 ATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLRR-SYGKEIDIWSAGVILYILLSGVPPFW 281
Query: 234 AKEHSDTYRRII--QVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQN 291
A+ + +I ++D P P +S +AKDL+ +ML KD R+ ++LEHPWI
Sbjct: 282 AENEKGIFDEVIKGEIDFVSEPWPSISESAKDLVRKMLTKDPKRRITAAQVLEHPWIKGG 341
Query: 292 AEP 294
P
Sbjct: 342 EAP 344
>At5g19360 calcium-dependent protein kinase - like
Length = 523
Score = 170 bits (431), Expect = 8e-43
Identities = 97/269 (36%), Positives = 154/269 (57%), Gaps = 9/269 (3%)
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
+ + + +GK LGRG+FG +L +K + A K + K +L + +RREV+I HL
Sbjct: 64 VKSSYTLGKELGRGQFGVTHLCTQKATGLQFACKTIAKRKLVNKEDIEDVRREVQIMHHL 123
Query: 93 R-HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYC 151
P+I+ L G + D+ V+L++E GEL+ + ++SER AA+ + ++ + + C
Sbjct: 124 TGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKGHYSERAAASLLRTIVQIIHTC 183
Query: 152 HGKHVIHRDIKPENLLIGAQGE---LKIADFGWSVHTFNRR--RTMCGTLDYLPPEMVES 206
H VIHRD+KPEN L+ ++ E LK DFG SV + + G+ Y+ PE++
Sbjct: 184 HSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKPGEVFKDIVGSAYYIAPEVLRR 243
Query: 207 VEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRII--QVDLKFPPKPIVSSAAKDL 264
++ DIWS+GV+ Y L GVPPF A+ + + I+ QVD P P++S AKDL
Sbjct: 244 -KYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILSGQVDFSSDPWPVISPQAKDL 302
Query: 265 ISQMLVKDSSERLPLHKLLEHPWIVQNAE 293
+ +ML D +RL ++L HPWI ++ E
Sbjct: 303 VRKMLNSDPKQRLTAAQVLNHPWIKEDGE 331
>At5g12180 calcium-dependent protein kinase
Length = 528
Score = 170 bits (430), Expect = 1e-42
Identities = 97/265 (36%), Positives = 152/265 (56%), Gaps = 9/265 (3%)
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR-HP 95
+ +GK LGRG+FG +L +K + H A K + K +L + +RREV+I HL P
Sbjct: 73 YSLGKELGRGQFGVTHLCTQKATGHQFACKTIAKRKLVNKEDIEDVRREVQIMHHLTGQP 132
Query: 96 HILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKH 155
+I+ L G + D+ V+L++E GEL+ + ++SER AA+ + ++ + + CH
Sbjct: 133 NIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKGHYSERAAASLLRTIVQIVHTCHSMG 192
Query: 156 VIHRDIKPENLLIGAQGE---LKIADFGWSVHTFNRR--RTMCGTLDYLPPEMVESVEHD 210
VIHRD+KPEN L+ + E LK DFG SV + + G+ Y+ PE+++ ++
Sbjct: 193 VIHRDLKPENFLLLNKDENSPLKATDFGLSVFYKPGEVFKDIVGSAYYIAPEVLKR-KYG 251
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQ--VDLKFPPKPIVSSAAKDLISQM 268
DIWS+GV+ Y L GVPPF A+ + + I++ VD P P +S AKDL+ +M
Sbjct: 252 PEADIWSIGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPQAKDLVKKM 311
Query: 269 LVKDSSERLPLHKLLEHPWIVQNAE 293
L D +RL ++L HPWI ++ E
Sbjct: 312 LNSDPKQRLTAAQVLNHPWIKEDGE 336
>At5g39440 serine/threonine-specific protein kinase -like protein
Length = 494
Score = 169 bits (427), Expect = 2e-42
Identities = 88/257 (34%), Positives = 145/257 (56%), Gaps = 4/257 (1%)
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
IL ++ IGK LG G F V LA + H VA+K+L +S+++ +E +++RE++I L
Sbjct: 15 ILPNYRIGKTLGHGSFAKVKLALHVATGHKVAIKILNRSKIKNMGIEIKVQREIKILRFL 74
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
HPHI+R Y +Y+++EY GEL+ + + E A + + YCH
Sbjct: 75 MHPHIIRQYEVIETPNDIYVVMEYVKSGELFDYIVEKGKLQEDEARHLFQQIISGVEYCH 134
Query: 153 GKHVIHRDIKPENLLIGAQGELKIADFGWS--VHTFNRRRTMCGTLDYLPPEMVESVEHD 210
++HRD+KPEN+L+ +Q +KI DFG S +H + +T CG+ +Y PE++ +
Sbjct: 135 RNMIVHRDLKPENVLLDSQCNIKIVDFGLSNVMHDGHFLKTSCGSPNYAAPEVISGKPYG 194
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLV 270
VDIWS GV+ Y L G PF+ + + + +I + P +S A+DLI +ML+
Sbjct: 195 PDVDIWSCGVILYALLCGTLPFDDENIPNVFEKIKRGMYTLPNH--LSHFARDLIPRMLM 252
Query: 271 KDSSERLPLHKLLEHPW 287
D + R+ + ++ +HPW
Sbjct: 253 VDPTMRISITEIRQHPW 269
>At1g35670 calcium-dependent protein kinase
Length = 495
Score = 168 bits (426), Expect = 3e-42
Identities = 101/292 (34%), Positives = 160/292 (54%), Gaps = 16/292 (5%)
Query: 6 ETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVAL 65
ET+P P + +T + +D + +GK LG+G+FG YL EK+++ A
Sbjct: 2 ETKPNPRRPSNTVLPYQTPRLRDH-------YLLGKKLGQGQFGTTYLCTEKSTSANYAC 54
Query: 66 KVLFKSQLQQSQVEHQLRREVEIQSHL-RHPHILRLYGYFYDQKRVYLILEYAPKGELYK 124
K + K +L + + RE++I HL HP+++R+ G + D V++++E GEL+
Sbjct: 55 KSIPKRKLVCREDYEDVWREIQIMHHLSEHPNVVRIKGTYEDSVFVHIVMEVCEGGELFD 114
Query: 125 ELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGA---QGELKIADFGW 181
+ +FSER A + ++ + CH V+HRD+KPEN L + +LK DFG
Sbjct: 115 RIVSKGHFSEREAVKLIKTILGVVEACHSLGVMHRDLKPENFLFDSPKDDAKLKATDFGL 174
Query: 182 SV--HTFNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSD 239
SV + G+ Y+ PE+++ + +D+WS GV+ Y L GVPPF A+ S
Sbjct: 175 SVFYKPGQYLYDVVGSPYYVAPEVLKKC-YGPEIDVWSAGVILYILLSGVPPFWAETESG 233
Query: 240 TYRRIIQ--VDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIV 289
+R+I+Q +D K P P +S AAKDLI +ML + +R+ H+ L HPWIV
Sbjct: 234 IFRQILQGKLDFKSDPWPTISEAAKDLIYKMLERSPKKRISAHEALCHPWIV 285
>At3g57530 calcium-dependent protein kinase
Length = 538
Score = 168 bits (425), Expect = 4e-42
Identities = 101/271 (37%), Positives = 154/271 (56%), Gaps = 10/271 (3%)
Query: 31 RWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQS 90
R I + + +G+ LGRG+FG YL +K ++ + A K + K +L+ + +RREVEI
Sbjct: 57 REIESKYTLGRELGRGEFGVTYLCTDKETDDVFACKSILKKKLRTAVDIEDVRREVEIMR 116
Query: 91 HL-RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALI 149
H+ HP+++ L + D+ V+L++E GEL+ + +++ER AA ++ +
Sbjct: 117 HMPEHPNVVTLKETYEDEHAVHLVMELCEGGELFDRIVARGHYTERAAAAVTKTIMEVVQ 176
Query: 150 YCHGKHVIHRDIKPENLLIGAQGE---LKIADFGWSV--HTFNRRRTMCGTLDYLPPEMV 204
CH V+HRD+KPEN L G + E LK DFG SV R + G+ Y+ PE++
Sbjct: 177 VCHKHGVMHRDLKPENFLFGNKKETAPLKAIDFGLSVFFKPGERFNEIVGSPYYMAPEVL 236
Query: 205 ESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQ--VDLKFPPKPIVSSAAK 262
+ + VDIWS GV+ Y L GVPPF A+ + II+ +D + P P VS AK
Sbjct: 237 KR-NYGPEVDIWSAGVILYILLCGVPPFWAETEQGVAQAIIRSVLDFRRDPWPKVSENAK 295
Query: 263 DLISQMLVKDSSERLPLHKLLEHPWIVQNAE 293
DLI +ML D RL ++L+HPW+ QNA+
Sbjct: 296 DLIRKMLDPDQKRRLTAQQVLDHPWL-QNAK 325
>At1g61950 hypothetical protein
Length = 547
Score = 168 bits (425), Expect = 4e-42
Identities = 94/272 (34%), Positives = 156/272 (56%), Gaps = 10/272 (3%)
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
I + +G+ LGRG+FG Y+ E +S A K + K +L +++ +RRE++I +L
Sbjct: 94 IKEKYSLGRELGRGQFGITYICTEISSGKNFACKSILKRKLIRTKDREDVRREIQIMHYL 153
Query: 93 R-HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYC 151
P+I+ + G + D++ V+L++E GEL+ ++ K ++SE+ AA + S+ + + C
Sbjct: 154 SGQPNIVEIKGAYEDRQSVHLVMELCEGGELFDKITKRGHYSEKAAAEIIRSVVKVVQIC 213
Query: 152 HGKHVIHRDIKPENLLIGAQGE----LKIADFGWSVHTFNRR--RTMCGTLDYLPPEMVE 205
H VIHRD+KPEN L+ ++ E LK DFG SV + + G+ Y+ PE+++
Sbjct: 214 HFMGVIHRDLKPENFLLSSKDEASSMLKATDFGVSVFIEEGKVYEDIVGSAYYVAPEVLK 273
Query: 206 SVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRII--QVDLKFPPKPIVSSAAKD 263
+ ++DIWS GV+ Y L G PPF A+ + I+ ++D + P P +S +AKD
Sbjct: 274 R-NYGKAIDIWSAGVILYILLCGNPPFWAETDKGIFEEILRGEIDFESEPWPSISESAKD 332
Query: 264 LISQMLVKDSSERLPLHKLLEHPWIVQNAEPS 295
L+ ML D +R ++LEHPWI + E S
Sbjct: 333 LVRNMLKYDPKKRFTAAQVLEHPWIREGGEAS 364
>At3g23000 SNF1 related protein kinase (ATSRPK1)
Length = 429
Score = 167 bits (424), Expect = 5e-42
Identities = 102/301 (33%), Positives = 163/301 (53%), Gaps = 16/301 (5%)
Query: 6 ETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVAL 65
E+ PQP Q S A + +L +++G+ LG G F V+LAR S+ +VA+
Sbjct: 2 ESLPQPQNQ--------SSPATTPAKILLGKYELGRRLGSGSFAKVHLARSIESDELVAV 53
Query: 66 KVLFKSQLQQSQVEHQLRREVEIQSHLRH-PHILRLYGYFYDQKRVYLILEYAPKGELYK 124
K++ K + +S +E ++ RE++ LRH P+IL+++ + ++YL++E A GEL+
Sbjct: 54 KIIEKKKTIESGMEPRIIREIDAMRRLRHHPNILKIHEVMATKSKIYLVMELASGGELFS 113
Query: 125 ELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSV- 183
++ + E A Y LA AL + H V HRD+KP+NLL+ QG LK++DFG S
Sbjct: 114 KVLRRGRLPESTARRYFQQLASALRFSHQDGVAHRDVKPQNLLLDEQGNLKVSDFGLSAL 173
Query: 184 --HTFN-RRRTMCGTLDYLPPEMVESVEHD-ASVDIWSLGVLCYEFLYGVPPFEAKEHSD 239
H N T CGT Y PE++ +D A D WS GV+ + L G PF+ +
Sbjct: 174 PEHLQNGLLHTACGTPAYTAPEVISRRGYDGAKADAWSCGVILFVLLVGDVPFDDSNIAA 233
Query: 240 TYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAEPSGIYR 299
YR+I + D +FP +S AK +I QML + R+ + +++ W ++ E S +R
Sbjct: 234 MYRKIHRRDYRFP--SWISKQAKSIIYQMLDPNPVTRMSIETVMKTNWFKKSLETSEFHR 291
Query: 300 S 300
+
Sbjct: 292 N 292
>At1g50700 hypothetical protein
Length = 521
Score = 167 bits (424), Expect = 5e-42
Identities = 92/267 (34%), Positives = 151/267 (56%), Gaps = 9/267 (3%)
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR-HP 95
+ + K LGRG+FG YL EK++ A K + K +L + +RRE++I HL P
Sbjct: 73 YTLSKELGRGQFGVTYLCTEKSTGKRFACKSISKKKLVTKGDKEDMRREIQIMQHLSGQP 132
Query: 96 HILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKH 155
+I+ G + D+K V L++E GEL+ + ++SER AA+ + + CH
Sbjct: 133 NIVEFKGAYEDEKAVNLVMELCAGGELFDRILAKGHYSERAAASVCRQIVNVVNICHFMG 192
Query: 156 VIHRDIKPENLLIGAQGE---LKIADFGWSVHTFNRR--RTMCGTLDYLPPEMVESVEHD 210
V+HRD+KPEN L+ ++ E +K DFG SV R + + G+ Y+ PE+++ +
Sbjct: 193 VMHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGRVYKDIVGSAYYVAPEVLKR-RYG 251
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRII--QVDLKFPPKPIVSSAAKDLISQM 268
+DIWS G++ Y L GVPPF A+ + I+ ++D + P P +S++AKDL+ +M
Sbjct: 252 KEIDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGEIDFESQPWPSISNSAKDLVRRM 311
Query: 269 LVKDSSERLPLHKLLEHPWIVQNAEPS 295
L +D R+ ++L+HPW+ + E S
Sbjct: 312 LTQDPKRRISAAEVLKHPWLREGGEAS 338
>At3g20410 calmodulin-domain protein kinase CDPK isoform 9
Length = 541
Score = 167 bits (422), Expect = 8e-42
Identities = 93/267 (34%), Positives = 148/267 (54%), Gaps = 9/267 (3%)
Query: 37 FDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR-HP 95
+ +GK LGRG+FG YL E ++ A K + K +L + +RRE++I HL P
Sbjct: 91 YTLGKELGRGQFGVTYLCTENSTGKKYACKSISKKKLVTKADKDDMRREIQIMQHLSGQP 150
Query: 96 HILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKH 155
+I+ G + D+K V L++E GEL+ + +++ER AA+ + + CH
Sbjct: 151 NIVEFKGAYEDEKAVNLVMELCAGGELFDRIIAKGHYTERAAASVCRQIVNVVKICHFMG 210
Query: 156 VIHRDIKPENLLIGAQGE---LKIADFGWSVHTFNRR--RTMCGTLDYLPPEMVESVEHD 210
V+HRD+KPEN L+ ++ E +K DFG SV + R + G+ Y+ PE++ +
Sbjct: 211 VLHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLRR-RYG 269
Query: 211 ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQ--VDLKFPPKPIVSSAAKDLISQM 268
VDIWS G++ Y L GVPPF A+ + I++ +D + P P +SS+AKDL+ +M
Sbjct: 270 KEVDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGHIDFESQPWPSISSSAKDLVRRM 329
Query: 269 LVKDSSERLPLHKLLEHPWIVQNAEPS 295
L D R+ +L+HPW+ + E S
Sbjct: 330 LTADPKRRISAADVLQHPWLREGGEAS 356
>At3g01090 putative SNF1-related protein kinase
Length = 512
Score = 167 bits (422), Expect = 8e-42
Identities = 88/258 (34%), Positives = 147/258 (56%), Gaps = 5/258 (1%)
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
IL ++ +G+ LG G FG V +A + H VA+K+L + +++ ++E ++RRE++I
Sbjct: 15 ILPNYKLGRTLGIGSFGRVKIAEHALTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLF 74
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
HPHI+RLY +YL++EY GEL+ + + E A + + + YCH
Sbjct: 75 MHPHIIRLYEVIETPTDIYLVMEYVNSGELFDYIVEKGRLQEDEARNFFQQIISGVEYCH 134
Query: 153 GKHVIHRDIKPENLLIGAQGELKIADFGWS--VHTFNRRRTMCGTLDYLPPEMVE-SVEH 209
V+HRD+KPENLL+ ++ +KIADFG S + + +T CG+ +Y PE++ +
Sbjct: 135 RNMVVHRDLKPENLLLDSKCNVKIADFGLSNIMRDGHFLKTSCGSPNYAAPEVISGKLYA 194
Query: 210 DASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQML 269
VD+WS GV+ Y L G PF+ + + +++I P +S A+DLI +ML
Sbjct: 195 GPEVDVWSCGVILYALLCGTLPFDDENIPNLFKKIKGGIYTLPSH--LSPGARDLIPRML 252
Query: 270 VKDSSERLPLHKLLEHPW 287
V D +R+ + ++ +HPW
Sbjct: 253 VVDPMKRVTIPEIRQHPW 270
>At5g45820 CBL-interacting protein kinase 20 (CIPK20)
Length = 439
Score = 165 bits (418), Expect = 2e-41
Identities = 90/265 (33%), Positives = 151/265 (56%), Gaps = 17/265 (6%)
Query: 33 ILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL 92
++ +++G+ LG+G F VY AR + VA+KV+ K ++ + + Q++RE+ + +
Sbjct: 8 LMRKYELGRLLGQGTFAKVYHARNIKTGESVAIKVIDKQKVAKVGLIDQIKREISVMRLV 67
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
RHPH++ L+ + ++Y +EY GEL+ ++ K K E A Y L A+ YCH
Sbjct: 68 RHPHVVFLHEVMASKTKIYFAMEYVKGGELFDKVSKGK-LKENIARKYFQQLIGAIDYCH 126
Query: 153 GKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRR-----TMCGTLDYLPPEMVESV 207
+ V HRD+KPENLL+ G+LKI+DFG S ++++ T CGT Y+ PE++
Sbjct: 127 SRGVYHRDLKPENLLLDENGDLKISDFGLSALRESKQQDGLLHTTCGTPAYVAPEVIGKK 186
Query: 208 EHD-ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLK----FPPKPIVSSAAK 262
+D A D+WS GV+ Y L G PF + + YR+I + + K FPP+ K
Sbjct: 187 GYDGAKADVWSCGVVLYVLLAGFLPFHEQNLVEMYRKITKGEFKCPNWFPPE------VK 240
Query: 263 DLISQMLVKDSSERLPLHKLLEHPW 287
L+S++L + + R+ + K++E+ W
Sbjct: 241 KLLSRILDPNPNSRIKIEKIMENSW 265
>At4g09570 calmodulin-domain protein kinase CDPK isoform 4 (CPK4)
Length = 501
Score = 165 bits (418), Expect = 2e-41
Identities = 98/265 (36%), Positives = 151/265 (56%), Gaps = 10/265 (3%)
Query: 39 IGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHL-RHPHI 97
+GK LG+G+FG YL EK+S+ A K + K +L + + RE++I HL HP++
Sbjct: 27 LGKKLGQGQFGTTYLCTEKSSSANYACKSIPKRKLVCREDYEDVWREIQIMHHLSEHPNV 86
Query: 98 LRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHVI 157
+R+ G + D V++++E GEL+ + FSER AA + ++ + CH V+
Sbjct: 87 VRIKGTYEDSVFVHIVMEVCEGGELFDRIVSKGCFSEREAAKLIKTILGVVEACHSLGVM 146
Query: 158 HRDIKPENLLIGA---QGELKIADFGWSV--HTFNRRRTMCGTLDYLPPEMVESVEHDAS 212
HRD+KPEN L + +LK DFG SV + G+ Y+ PE+++ +
Sbjct: 147 HRDLKPENFLFDSPSDDAKLKATDFGLSVFYKPGQYLYDVVGSPYYVAPEVLKKC-YGPE 205
Query: 213 VDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQ--VDLKFPPKPIVSSAAKDLISQMLV 270
+D+WS GV+ Y L GVPPF A+ S +R+I+Q +D K P P +S AKDLI +ML
Sbjct: 206 IDVWSAGVILYILLSGVPPFWAETESGIFRQILQGKIDFKSDPWPTISEGAKDLIYKMLD 265
Query: 271 KDSSERLPLHKLLEHPWIV-QNAEP 294
+ +R+ H+ L HPWIV ++A P
Sbjct: 266 RSPKKRISAHEALCHPWIVDEHAAP 290
>At1g30270 serine/threonine kinase, putative
Length = 482
Score = 165 bits (418), Expect = 2e-41
Identities = 88/274 (32%), Positives = 153/274 (55%), Gaps = 9/274 (3%)
Query: 20 SEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVE 79
S SGS+ R + +++G+ LG G F V AR + VA+KV+ K ++ ++++
Sbjct: 15 STPSGSSSGGRTRV-GKYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMI 73
Query: 80 HQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAAT 139
Q++RE+ ++HP+++R++ + ++Y +LE+ GEL+ ++ E A
Sbjct: 74 AQIKREISTMKLIKHPNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDEARK 133
Query: 140 YVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWS-----VHTFNRRRTMCG 194
Y L A+ YCH + V HRD+KPENLL+ A G LK++DFG S V T CG
Sbjct: 134 YFQQLINAVDYCHSRGVYHRDLKPENLLLDANGALKVSDFGLSALPQQVREDGLLHTTCG 193
Query: 195 TLDYLPPEMVESVEHD-ASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPP 253
T +Y+ PE++ + +D A D+WS GV+ + + G PFE + Y++I + + P
Sbjct: 194 TPNYVAPEVINNKGYDGAKADLWSCGVILFVLMAGYLPFEDSNLTSLYKKIFKAEFTCP- 252
Query: 254 KPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPW 287
P S++AK LI ++L + + R+ +++E+ W
Sbjct: 253 -PWFSASAKKLIKRILDPNPATRITFAEVIENEW 285
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,894,038
Number of Sequences: 26719
Number of extensions: 288985
Number of successful extensions: 3668
Number of sequences better than 10.0: 1003
Number of HSP's better than 10.0 without gapping: 923
Number of HSP's successfully gapped in prelim test: 80
Number of HSP's that attempted gapping in prelim test: 690
Number of HSP's gapped (non-prelim): 1111
length of query: 300
length of database: 11,318,596
effective HSP length: 99
effective length of query: 201
effective length of database: 8,673,415
effective search space: 1743356415
effective search space used: 1743356415
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138017.6


