

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138017.12 + phase: 1 /pseudo
(95 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
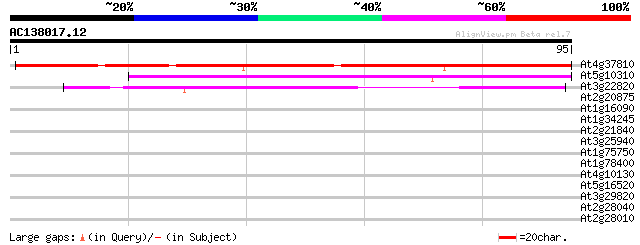
Score E
Sequences producing significant alignments: (bits) Value
At4g37810 hypothetical protein 87 1e-18
At5g10310 putative protein 68 7e-13
At3g22820 unknown protein 45 5e-06
At2g20875 putative protein 30 0.27
At1g16090 unknown protein 29 0.35
At1g34245 unknown protein 28 0.60
At2g21840 unknown protein 28 0.78
At3g25940 unknown protein 27 2.3
At1g75750 unknown protein 26 3.9
At1g78400 similar to exopolygalacturonase precursor sp|Q00293|PG... 25 5.1
At4g10130 putative protein 25 6.6
At5g16520 unknown protein 25 8.7
At3g29820 hypothetical protein 25 8.7
At2g28040 chloroplast nucleoid DNA binding like protein 25 8.7
At2g28010 putative chloroplast nucleoid DNA binding protein 25 8.7
>At4g37810 hypothetical protein
Length = 128
Score = 87.0 bits (214), Expect = 1e-18
Identities = 49/103 (47%), Positives = 66/103 (63%), Gaps = 12/103 (11%)
Query: 2 GRPISNLNEVAKNGMEENGSMLAEKLQIGSRPPKCER-RCRSCVHCEAVQVPIVPSKVQI 60
GRP + E K+G +++ M+ L IGSRPP+CER RCRSC HCEA+QVP P + ++
Sbjct: 28 GRPEPDSVEFTKSG-DQDVKMMMRGL-IGSRPPRCERVRCRSCGHCEAIQVPTNP-QTKL 84
Query: 61 HRSHYDSAAYSS--------RGDGLSNYKPISWKCKCGDYFFN 95
H S++ SS RGD +NYKP+SWKCKCG+ +N
Sbjct: 85 HSPLTTSSSSSSETIHLDYTRGDDSTNYKPMSWKCKCGNSIYN 127
>At5g10310 putative protein
Length = 122
Score = 68.2 bits (165), Expect = 7e-13
Identities = 29/82 (35%), Positives = 48/82 (58%), Gaps = 7/82 (8%)
Query: 21 SMLAEKLQIGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDSAAY-------SSR 73
+++ +K ++GS PP C RC +C C A+QVP +P++ + R + S + ++
Sbjct: 40 ALIEDKARLGSTPPSCHNRCNNCHPCMAIQVPTLPTRSRFTRVNPFSGGFVRPPSSLTTV 99
Query: 74 GDGLSNYKPISWKCKCGDYFFN 95
D SNYKP+ WKC C +F+N
Sbjct: 100 LDQYSNYKPMGWKCHCNGHFYN 121
>At3g22820 unknown protein
Length = 107
Score = 45.4 bits (106), Expect = 5e-06
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 21/87 (24%)
Query: 10 EVAKNGMEENGSMLAEKLQ--IGSRPPKCERRCRSCVHCEAVQVPIVPSKVQIHRSHYDS 67
E+A++G+ G ++ +K GS PP C +C C C+AV VPI P +
Sbjct: 38 EIARSGLP--GQIVDQKRLGGPGSVPPMCRLKCGKCEPCKAVHVPIQPGLIM-------- 87
Query: 68 AAYSSRGDGLSNYKPISWKCKCGDYFF 94
Y P +W+CKCG+ F
Sbjct: 88 ---------PLEYYPEAWRCKCGNKLF 105
>At2g20875 putative protein
Length = 104
Score = 29.6 bits (65), Expect = 0.27
Identities = 13/30 (43%), Positives = 15/30 (49%)
Query: 30 GSRPPKCERRCRSCVHCEAVQVPIVPSKVQ 59
GSR P C C SC C V V V + V+
Sbjct: 54 GSRLPDCSHACGSCSPCRLVMVSFVCASVE 83
>At1g16090 unknown protein
Length = 382
Score = 29.3 bits (64), Expect = 0.35
Identities = 23/76 (30%), Positives = 31/76 (40%), Gaps = 9/76 (11%)
Query: 5 ISNLNEVAKNGMEENGSMLAEKLQIGSRPPKCE------RRCRSCVHCEAVQVPIVPSKV 58
++ N + G N SM +KLQIG C+ R RSC Q I+ ++
Sbjct: 131 LTGRNTLVAVGCNNNASMTDDKLQIGGCESTCDVGFGQRGRNRSCNGYRCCQAKILSDRL 190
Query: 59 Q---IHRSHYDSAAYS 71
Q I D AYS
Sbjct: 191 QQIGIKIESLDDEAYS 206
>At1g34245 unknown protein
Length = 120
Score = 28.5 bits (62), Expect = 0.60
Identities = 12/39 (30%), Positives = 18/39 (45%)
Query: 13 KNGMEENGSMLAEKLQIGSRPPKCERRCRSCVHCEAVQV 51
K + +NG + E GS P C C +C C+ V +
Sbjct: 53 KKEISKNGGVEMEMYPTGSSLPDCSYACGACSPCKRVMI 91
>At2g21840 unknown protein
Length = 746
Score = 28.1 bits (61), Expect = 0.78
Identities = 18/54 (33%), Positives = 25/54 (45%), Gaps = 11/54 (20%)
Query: 22 MLAEKLQIGSRPPKCERR----------CRSCVHCEAVQVPIVPSKVQIHRSHY 65
MLA L+ GS+ C+ C H E ++ IVP K++ H SHY
Sbjct: 581 MLAISLKSGSKCTACQLNITKYGYHCTICEINFHIECIKAVIVPRKIKSH-SHY 633
>At3g25940 unknown protein
Length = 119
Score = 26.6 bits (57), Expect = 2.3
Identities = 11/29 (37%), Positives = 15/29 (50%)
Query: 21 SMLAEKLQIGSRPPKCERRCRSCVHCEAV 49
S+ EK Q + PK ++ C C H E V
Sbjct: 62 SLFGEKTQAEAELPKIKKACEKCQHPELV 90
>At1g75750 unknown protein
Length = 98
Score = 25.8 bits (55), Expect = 3.9
Identities = 14/45 (31%), Positives = 20/45 (44%), Gaps = 4/45 (8%)
Query: 9 NEVAKNGMEEN---GSMLAEKLQIGSRPPKCERRCRSCVH-CEAV 49
N KNG + GS + ++ RP C R C +C + C V
Sbjct: 27 NSQKKNGYAKKIDCGSACVARCRLSRRPRLCHRACGTCCYRCNCV 71
>At1g78400 similar to exopolygalacturonase precursor
sp|Q00293|PGLX_ASPTU
Length = 412
Score = 25.4 bits (54), Expect = 5.1
Identities = 13/45 (28%), Positives = 21/45 (45%), Gaps = 3/45 (6%)
Query: 48 AVQVPIVPSKVQIHRSHYDSAAYSSRGDGLSNYKPI---SWKCKC 89
A + PS + R +D +Y +RGDG ++ +WK C
Sbjct: 18 ASSISAAPSAALVGRKVFDVRSYGARGDGKTDNTMAFTKAWKDAC 62
>At4g10130 putative protein
Length = 174
Score = 25.0 bits (53), Expect = 6.6
Identities = 13/44 (29%), Positives = 21/44 (47%), Gaps = 15/44 (34%)
Query: 65 YDSAAYSSRGDGLS---------------NYKPISWKCKCGDYF 93
YD+ SSR DG++ + + ++C+CGDYF
Sbjct: 75 YDNDLRSSRHDGITADEISIEDMSVEITGDVIDLFYQCRCGDYF 118
>At5g16520 unknown protein
Length = 608
Score = 24.6 bits (52), Expect = 8.7
Identities = 10/36 (27%), Positives = 19/36 (52%)
Query: 49 VQVPIVPSKVQIHRSHYDSAAYSSRGDGLSNYKPIS 84
++ ++ + S +DSA+ + GD SN K I+
Sbjct: 496 IEKNVISQPASLELSSFDSASSLAHGDNFSNKKSIT 531
>At3g29820 hypothetical protein
Length = 332
Score = 24.6 bits (52), Expect = 8.7
Identities = 7/14 (50%), Positives = 10/14 (71%)
Query: 76 GLSNYKPISWKCKC 89
G+ +Y P+SWK C
Sbjct: 278 GICDYHPLSWKANC 291
>At2g28040 chloroplast nucleoid DNA binding like protein
Length = 395
Score = 24.6 bits (52), Expect = 8.7
Identities = 17/44 (38%), Positives = 20/44 (44%), Gaps = 12/44 (27%)
Query: 26 KLQIGSRPPKCER-----------RCRSCVHCEAVQVPIV-PSK 57
KLQIG+ P + E +C CVHC PI PSK
Sbjct: 68 KLQIGTPPFEIEAVLDTGSEHIWTQCLPCVHCYNQTAPIFDPSK 111
>At2g28010 putative chloroplast nucleoid DNA binding protein
Length = 396
Score = 24.6 bits (52), Expect = 8.7
Identities = 16/51 (31%), Positives = 23/51 (44%), Gaps = 12/51 (23%)
Query: 19 NGSMLAEKLQIGSRPPKCER-----------RCRSCVHCEAVQVPIV-PSK 57
+ S+ KLQ+G+ P + + +C CVHC PI PSK
Sbjct: 61 DNSVYLMKLQVGTPPFEIQAIIDTGSEITWTQCLPCVHCYEQNAPIFDPSK 111
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,458,527
Number of Sequences: 26719
Number of extensions: 89958
Number of successful extensions: 204
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 191
Number of HSP's gapped (non-prelim): 15
length of query: 95
length of database: 11,318,596
effective HSP length: 71
effective length of query: 24
effective length of database: 9,421,547
effective search space: 226117128
effective search space used: 226117128
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC138017.12


