

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138015.15 + phase: 0 /pseudo
(821 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
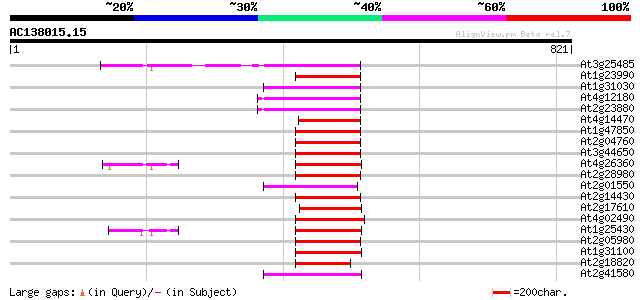
Score E
Sequences producing significant alignments: (bits) Value
At3g25485 unknown protein 97 4e-20
At1g23990 reverse transcriptase, putative 97 4e-20
At1g31030 F17F8.5 92 2e-18
At4g12180 putative reverse transcriptase 89 1e-17
At2g23880 putative non-LTR retroelement reverse transcriptase 89 1e-17
At4g14470 reverse transcriptase like protein 85 1e-16
At1g47850 hypothetical protein 84 3e-16
At2g04760 putative non-LTR retroelement reverse transcriptase 83 7e-16
At3g44650 putative protein 82 9e-16
At4g26360 putative protein 82 2e-15
At2g28980 putative non-LTR retroelement reverse transcriptase 81 2e-15
At2g01550 putative non-LTR retroelement reverse transcriptase 80 4e-15
At2g14430 putative non-LTR retroelement reverse transcriptase 80 6e-15
At2g17610 putative non-LTR retroelement reverse transcriptase 78 2e-14
At4g02490 77 3e-14
At1g25430 hypothetical protein 77 3e-14
At2g05980 putative non-LTR retroelement reverse transcriptase 76 7e-14
At1g31100 hypothetical protein 76 9e-14
At2g18820 putative non-LTR retroelement reverse transcriptase 75 1e-13
At2g41580 putative non-LTR retroelement reverse transcriptase 74 4e-13
>At3g25485 unknown protein
Length = 979
Score = 97.1 bits (240), Expect = 4e-20
Identities = 86/384 (22%), Positives = 160/384 (41%), Gaps = 47/384 (12%)
Query: 134 LHTNLLTNIWFKLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLIE 193
+ T+ + KLKALK +++L K+ ++A L Q + P +IE
Sbjct: 235 MSTSTMFRFTKKLKALKPKIRQLAKEKMGNLVKRTKEAHDTLCQKQEENLKSPSPQAMIE 294
Query: 194 AEKVCLSSLKKW---STIEEQIWI*KSRANWIQLGDSTQNFSMPLQRKEDVRISLSSS*Q 250
+ + +KW + IEE+ KS+ +W+Q+GD D + Q
Sbjct: 295 ETE----AHEKWELLAEIEEKYLKQKSKLHWMQVGDKNNKTFHRALVPNDFEGATLEELQ 350
Query: 251 KMAPKLTSITSSRRRLEGSI*N*WVVLLIRYLWWTKISSREVLCYLSISKTCCAQSSQPW 310
+ P + R +E ++ E+ L + + +
Sbjct: 351 VLLPFRCTAEDRSRLME------------------PVTGEEIKTVLFFMSSDKSPGPDGF 392
Query: 311 RSKMRYSPWILLRLQ-VLMVTVFVSLNALGTLLVIVSLMLY*NSLRLVSCLILSTILM*L 369
++ + W ++ + ++ V F L + V+ IL+ I
Sbjct: 393 TTEFYKATWEIIGAEFIVAVKSFFEKGFLP---------------KGVNSTILALI---- 433
Query: 370 CYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKGRVIFDNI 429
PKK +++ P+ ++ K KE + FVK R++ N+
Sbjct: 434 --PKKFETKEMKDYRPISCCNVIYKVISKLIAKRLKEILPQFIAGNQSAFVKDRLLIQNL 491
Query: 430 ILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN*AMACLTT 489
+L+ E+VK Y ++ +S RC +KID+ KA+ SV+ F+ ++ + F +F++ M C+TT
Sbjct: 492 LLATEIVKDYHKESVSDRCAIKIDISKAFDSVQWSFLENVLHSLNFSQEFIHWIMLCITT 551
Query: 490 TSYTYNVNGDLTRPFAAKKGLRQG 513
S++ VNG+L F + +GLRQG
Sbjct: 552 ASFSVQVNGELVGFFNSSRGLRQG 575
>At1g23990 reverse transcriptase, putative
Length = 657
Score = 97.1 bits (240), Expect = 4e-20
Identities = 44/95 (46%), Positives = 68/95 (71%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y ++ +S RC +KID+ KA+ SV+ FI ++L M FP +
Sbjct: 14 FVKDRLLIENLLLATELVKDYHKESVSSRCAIKIDISKAFNSVQWSFIRNILLSMDFPME 73
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
FV+ M C++T S++ VNG+L F +K+GLRQG
Sbjct: 74 FVHWIMLCISTASFSVQVNGELVGFFQSKRGLRQG 108
>At1g31030 F17F8.5
Length = 872
Score = 91.7 bits (226), Expect = 2e-18
Identities = 50/142 (35%), Positives = 78/142 (54%)
Query: 372 PKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKGRVIFDNIIL 431
PKKL ++R P+ ++ K K + + FVK R++ +N++L
Sbjct: 165 PKKLAAKEMRDYRPISCCNVLYKVISKIIANRLKLLLPRFIAENQSAFVKDRLLIENLLL 224
Query: 432 SHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN*AMACLTTTS 491
+ ELVK Y + IS RC +KID+ KA+ SV+ F+ ++ M F F++ C+TT S
Sbjct: 225 ATELVKDYHKDSISARCAIKIDISKAFDSVQWSFLTNTLVAMNFSPTFIHWINLCITTAS 284
Query: 492 YTYNVNGDLTRPFAAKKGLRQG 513
++ VNGDL F +K+GLRQG
Sbjct: 285 FSVQVNGDLVGYFQSKRGLRQG 306
>At4g12180 putative reverse transcriptase
Length = 662
Score = 88.6 bits (218), Expect = 1e-17
Identities = 50/151 (33%), Positives = 84/151 (55%), Gaps = 2/151 (1%)
Query: 363 STILM*LCYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKG 422
STIL + PKK+ +++ P+ ++ K K + F+K
Sbjct: 29 STILALI--PKKMEAKEIKDYRPISCCNVLYKVISKIIANRLKRVLPQFIAGNQSAFIKD 86
Query: 423 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN* 482
R++ +N++L+ ELVK Y + +S RC +KID+ KA+ SV+ F+ ++L + FP +FV+
Sbjct: 87 RLLIENLLLATELVKDYHKDSVSERCAIKIDISKAFDSVQWSFLRNVLLTLDFPQEFVHW 146
Query: 483 AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
M C+TT S++ VN +L F + +GLRQG
Sbjct: 147 IMLCVTTASFSVQVNRELAGYFNSLRGLRQG 177
>At2g23880 putative non-LTR retroelement reverse transcriptase
Length = 1216
Score = 88.6 bits (218), Expect = 1e-17
Identities = 50/151 (33%), Positives = 83/151 (54%), Gaps = 2/151 (1%)
Query: 363 STILM*LCYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKG 422
STIL + PKK +++ P+ + K K +V FVK
Sbjct: 232 STILALI--PKKKEAREIKDYRPISCCNVLYKAISKILANRLKRILPKFIVGNQSAFVKD 289
Query: 423 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN* 482
R++ +N++L+ ELVK Y + IS RC +KID+ KA+ S++ F+ +++ M FP +F++
Sbjct: 290 RLLIENVLLATELVKDYHKDSISTRCAMKIDISKAFDSLQWSFLTHVLAAMNFPGEFIHW 349
Query: 483 AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
C++T S++ VNG+L F + +GLRQG
Sbjct: 350 ISLCMSTASFSIQVNGELAGYFRSARGLRQG 380
>At4g14470 reverse transcriptase like protein
Length = 318
Score = 85.1 bits (209), Expect = 1e-16
Identities = 39/91 (42%), Positives = 62/91 (67%)
Query: 423 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN* 482
R++ +N++L+ ELVK Y + +S RC +KID+ KA+ SV+ F+ ++L + FP FV+
Sbjct: 5 RLLIENLLLATELVKDYHKDSVSERCAIKIDISKAFDSVQWTFLKNVLLTLDFPQVFVHW 64
Query: 483 AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
M C+TT S++ VNG+L F + +GLRQG
Sbjct: 65 IMLCVTTASFSVQVNGELAGYFNSSRGLRQG 95
>At1g47850 hypothetical protein
Length = 799
Score = 84.0 bits (206), Expect = 3e-16
Identities = 39/95 (41%), Positives = 63/95 (66%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FV+ R++ +N++L+ ELVK Y + ISPRC +KID+ KA+ SV+ F+ + + FP
Sbjct: 138 FVRERLLIENVLLATELVKDYHKDSISPRCAMKIDISKAFDSVQWQFLLNTLEALNFPEN 197
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F + C++T +++ VNG+L F +K+GLRQG
Sbjct: 198 FCHWIKLCISTATFSVQVNGELAGFFGSKRGLRQG 232
>At2g04760 putative non-LTR retroelement reverse transcriptase
Length = 631
Score = 82.8 bits (203), Expect = 7e-16
Identities = 40/95 (42%), Positives = 62/95 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y + ISPRC +KID+ KA+ SV+ F+ + + FP K
Sbjct: 135 FVKERLLMENVLLATELVKDYHKDSISPRCAMKIDISKAFDSVQWQFLLNTLEALNFPEK 194
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
+ C++T +++ VNG+L F K+GLRQG
Sbjct: 195 IRHWIKLCISTATFSVQVNGELAGFFGNKRGLRQG 229
>At3g44650 putative protein
Length = 762
Score = 82.4 bits (202), Expect = 9e-16
Identities = 40/95 (42%), Positives = 61/95 (64%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ +LVK Y + IS RC +KID+ KA SV+ F+ + M FP
Sbjct: 118 FVKDRLLIENVLLATDLVKDYHKDSISERCAIKIDISKASDSVQWSFLINTLTAMHFPEM 177
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F++ C+TT S++ VNG+L F + +GLRQG
Sbjct: 178 FIHWIRLCITTPSFSVQVNGELAGFFQSSRGLRQG 212
>At4g26360 putative protein
Length = 1141
Score = 81.6 bits (200), Expect = 2e-15
Identities = 41/96 (42%), Positives = 64/96 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+ GR++ +N++L+ E+V Y+ + IS R M+K+DL+KA+ SV FI +L +G P K
Sbjct: 537 FLPGRLLAENVLLATEMVHGYNWRNISLRGMLKVDLRKAFDSVRWEFIIAALLALGVPTK 596
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
F+N C++T ++T +VNG F + KGLRQGD
Sbjct: 597 FINWIHQCISTPTFTVSVNGCCGGFFKSAKGLRQGD 632
Score = 44.3 bits (103), Expect = 3e-04
Identities = 35/128 (27%), Positives = 59/128 (45%), Gaps = 24/128 (18%)
Query: 137 NLLTNIWF--------------KLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQL 182
NL+ ++W+ KLKALKK +K+ + ++ K+ E+A L+ Q L
Sbjct: 255 NLVWDVWYSTNVVGSSMFRVSKKLKALKKPIKDFSRLNYSNLEKRTEEAHETLLSFQ-NL 313
Query: 183 SSDPMNLDLIEAEKVCLSSLKKW---STIEEQIWI*KSRANWIQLGDSTQNFSMPLQRKE 239
+ D +L+ E L + +KW +T EE + +SR W GD + R
Sbjct: 314 TLDNPSLENAAHE---LEAQRKWQILATAEESFFRQRSRVTWFAEGDGNTRY---FHRMA 367
Query: 240 DVRISLSS 247
D R S+++
Sbjct: 368 DSRKSVNT 375
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 81.3 bits (199), Expect = 2e-15
Identities = 37/95 (38%), Positives = 63/95 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y ++ ++PRC +KID+ KA+ SV+ F+ + + FP
Sbjct: 862 FVKERLLMENVLLATELVKDYHKESVTPRCAMKIDISKAFDSVQWQFLLNTLEALNFPET 921
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F + C++T +++ VNG+L F + +GLRQG
Sbjct: 922 FRHWIKLCISTATFSVQVNGELAGFFGSSRGLRQG 956
Score = 36.2 bits (82), Expect = 0.076
Identities = 25/96 (26%), Positives = 42/96 (43%), Gaps = 1/96 (1%)
Query: 136 TNLLTNIWFKLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLIEAE 195
T+ L KLK LK L+EL K+ +A L + Q ++P + + I E
Sbjct: 578 TSALYRFSKKLKTLKPHLRELGKEKLGDLPKRTREAHILLCEKQATTLANP-SQETIAEE 636
Query: 196 KVCLSSLKKWSTIEEQIWI*KSRANWIQLGDSTQNF 231
+ S +EE KS+ +W+ +GD ++
Sbjct: 637 LKAYTDWTHLSELEEGFLKQKSKLHWMNVGDGNNSY 672
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 80.5 bits (197), Expect = 4e-15
Identities = 44/137 (32%), Positives = 74/137 (53%)
Query: 372 PKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKGRVIFDNIIL 431
PKKL +++ P+ ++ K K + FVK R++ +N++L
Sbjct: 933 PKKLEAKEMKDYRPISCCNVIYKVISKIIANRLKHVLPNFIAGNQSAFVKDRLLIENLLL 992
Query: 432 SHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN*AMACLTTTS 491
+ ELVK Y + IS RC +KID+ KA+ SV+ F+ ++ + FP +FV+ M C+TT S
Sbjct: 993 ATELVKDYHKDTISGRCAIKIDISKAFDSVQWSFLKNVLSALDFPPEFVHWVMLCVTTAS 1052
Query: 492 YTYNVNGDLTRPFAAKK 508
++ VNG+L F + +
Sbjct: 1053 FSVQVNGELAGYFQSSR 1069
Score = 35.4 bits (80), Expect = 0.13
Identities = 25/94 (26%), Positives = 45/94 (47%), Gaps = 1/94 (1%)
Query: 133 NLHTNLLTNIWFKLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLI 192
++ T+ L KLKALK L+ L K+ +A +L Q+ S +P + +
Sbjct: 693 HMSTSSLFRFTKKLKALKPKLRGLAKEKMGNLVKRTREAYLSLCQAQQSNSQNP-SQRAM 751
Query: 193 EAEKVCLSSLKKWSTIEEQIWI*KSRANWIQLGD 226
E E + ++IEE+ S+ +W+++GD
Sbjct: 752 EIESEAYVRWDRIASIEEKYLKQVSKLHWLKVGD 785
>At2g14430 putative non-LTR retroelement reverse transcriptase
Length = 1277
Score = 79.7 bits (195), Expect = 6e-15
Identities = 38/95 (40%), Positives = 62/95 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FV+ R++ +N++L+ ELVK Y + ISPRC +KID+ KA+ SV+ F+ + + FP K
Sbjct: 707 FVRERLLMENVLLATELVKDYHKDSISPRCAMKIDISKAFDSVQWQFLLNTLEALKFPEK 766
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F + C++T +++ VN + F +K+GLRQG
Sbjct: 767 FRHWIKLCISTATFSVQVNSEQAGFFGSKRGLRQG 801
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 77.8 bits (190), Expect = 2e-14
Identities = 38/92 (41%), Positives = 60/92 (64%), Gaps = 1/92 (1%)
Query: 424 VIFDNIILSHELVKS-YSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN* 482
+I DNI+++HEL+ S +++K + P K+D+ KA+ +E FI +M +MGF K+ N
Sbjct: 1 MITDNILIAHELIHSLHTKKLVQPFVATKLDITKAFDKIEWGFIEAIMKQMGFSEKWCNW 60
Query: 483 AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
M C+TTT+Y+ +NG R K+G+RQGD
Sbjct: 61 IMTCITTTTYSILINGQPVRRIIPKRGIRQGD 92
>At4g02490
Length = 657
Score = 77.4 bits (189), Expect = 3e-14
Identities = 38/102 (37%), Positives = 65/102 (63%), Gaps = 1/102 (0%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+KGR++ +N++L+ ELV ++ +G + R +++DL KAY +V F+ ++ + P
Sbjct: 106 FIKGRLLCENVLLASELVDNFQAEGDTSRGCLQVDLTKAYDNVNWEFLINILKALNLPPI 165
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD-LSLHI 519
F+N C++T SY+ NG+L F KKG+RQGD +S H+
Sbjct: 166 FINWIWVCISTPSYSIAYNGELIGFFVGKKGIRQGDPMSSHL 207
>At1g25430 hypothetical protein
Length = 1213
Score = 77.4 bits (189), Expect = 3e-14
Identities = 38/96 (39%), Positives = 61/96 (62%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+ GR + +N++L+ +LV Y+ ISPR M+K+DL+KA+ SV F+ + + P K
Sbjct: 559 FLPGRSLAENVLLATDLVHGYNWSNISPRGMLKVDLKKAFDSVRWEFVIAALRALAIPEK 618
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
F+N C++T ++T ++NG F + KGLRQGD
Sbjct: 619 FINWISQCISTPTFTVSINGGNGGFFKSTKGLRQGD 654
Score = 43.1 bits (100), Expect = 6e-04
Identities = 32/109 (29%), Positives = 52/109 (47%), Gaps = 16/109 (14%)
Query: 145 KLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDL---IEAEKVCLSS 201
KLKALKK +K+ + ++ K+ ++A L+ Q + +DP ++ +EAE
Sbjct: 285 KLKALKKPIKDFSRLNYSELEKRTKEAHDFLIGCQDRTLADPTPINASFELEAE------ 338
Query: 202 LKKW---STIEEQIWI*KSRANWIQLGDSTQNFSMPLQRKEDVRISLSS 247
+KW + EE + KSR +W GD + R D R S +S
Sbjct: 339 -RKWHILTAAEESFFRQKSRISWFAEGDGNTKY---FHRMADARNSSNS 383
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 76.3 bits (186), Expect = 7e-14
Identities = 36/95 (37%), Positives = 63/95 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+K R++ +N++L+ ELVK Y ++ IS R +KID+ KA+ V+ PF+ ++ + P
Sbjct: 712 FIKDRLMMENLLLASELVKDYHKESISSRSALKIDISKAFDFVQWPFLINVLKAIHLPEM 771
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F++ C+ T S++ VNG+L+ F +++GLRQG
Sbjct: 772 FIHWIELCIGTASFSVQVNGELSGFFRSERGLRQG 806
>At1g31100 hypothetical protein
Length = 1090
Score = 75.9 bits (185), Expect = 9e-14
Identities = 37/96 (38%), Positives = 63/96 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+ GR + +N++L+ ELV+ Y+R+ I PR M+K+DL+KA+ S+ FI + +G P +
Sbjct: 474 FLPGRFLAENVLLATELVQGYNRQNIDPRGMLKVDLRKAFDSIRWDFIISALKAIGIPDR 533
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
FV C++T +++ VNG+ F + +GLRQG+
Sbjct: 534 FVYWITQCISTPTFSVCVNGNTGGFFKSTRGLRQGN 569
Score = 40.4 bits (93), Expect = 0.004
Identities = 25/90 (27%), Positives = 45/90 (49%), Gaps = 7/90 (7%)
Query: 145 KLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLIEAEKVCLSSLKK 204
KLKALK ++ + +F K+V++A ++ Q + SDP I + + + +K
Sbjct: 196 KLKALKNPIRTFSMENFSNLEKRVKEAHNLVLYRQNKTLSDP----TIPNAALEMEAQRK 251
Query: 205 WSTI---EEQIWI*KSRANWIQLGDSTQNF 231
W + EE + +SR W+ GDS ++
Sbjct: 252 WLILVKAEESFFCQRSRVTWMGEGDSNTSY 281
>At2g18820 putative non-LTR retroelement reverse transcriptase
Length = 1412
Score = 75.5 bits (184), Expect = 1e-13
Identities = 33/80 (41%), Positives = 54/80 (67%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+ R++ +N++L+ ELVK Y + G+SPRC +KIDL KA+ SV+ PF+ + + P K
Sbjct: 831 FISDRLLMENLLLASELVKDYHKDGLSPRCAMKIDLSKAFDSVQWPFLLNTLAALDIPEK 890
Query: 479 FVN*AMACLTTTSYTYNVNG 498
F++ C++T S++ VNG
Sbjct: 891 FIHWINLCISTASFSVQVNG 910
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 73.6 bits (179), Expect = 4e-13
Identities = 47/145 (32%), Positives = 77/145 (52%), Gaps = 2/145 (1%)
Query: 372 PKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKGRVIFDNIIL 431
PK ++ + P+ + ++ K K+ V++ FV R++ DNIIL
Sbjct: 222 PKITKPARMADIRPISLCSVMYKIISKILSARLKKYLPVIVSPTQSAFVAERLVSDNIIL 281
Query: 432 SHELVKSY-SRKGISPRCMV-KIDLQKAYYSVE*PFINYLMLEMGFPCKFVN*AMACLTT 489
+HE+V + + + IS MV K D+ KAY VE PF+ ++L +GF ++N MAC+++
Sbjct: 282 AHEIVHNLRTNEKISKDFMVFKTDMSKAYDRVEWPFLKGILLALGFNSTWINWMMACVSS 341
Query: 490 TSYTYNVNGDLTRPFAAKKGLRQGD 514
SY+ +NG +GLRQGD
Sbjct: 342 VSYSVLINGQPFGHITPHRGLRQGD 366
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.348 0.153 0.523
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,864,524
Number of Sequences: 26719
Number of extensions: 597779
Number of successful extensions: 2349
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 2214
Number of HSP's gapped (non-prelim): 107
length of query: 821
length of database: 11,318,596
effective HSP length: 108
effective length of query: 713
effective length of database: 8,432,944
effective search space: 6012689072
effective search space used: 6012689072
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC138015.15


