

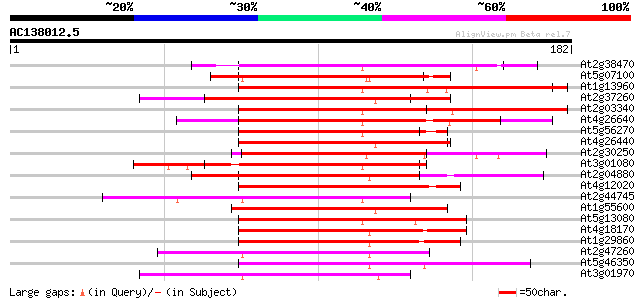
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138012.5 + phase: 0
(182 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g38470 putative WRKY-type DNA binding protein 110 4e-25
At5g07100 SPF1-like protein 97 5e-21
At1g13960 WRKY DNA-binding protein 4 (WRKY4) 96 1e-20
At2g37260 putative WRKY-type DNA binding protein 95 2e-20
At2g03340 putative WRKY DNA-binding protein 94 3e-20
At4g26640 WRKY transcription factor 20 (WRKY20) 93 7e-20
At5g56270 transcription factor NtWRKY4-like 93 9e-20
At4g26440 putative protein 89 1e-18
At2g30250 putative WRKY-type DNA binding protein 89 2e-18
At3g01080 WRKY transcription factor 58 (WRKY58) 88 2e-18
At2g04880 WRKY transcription factor 1 splice variant 1 (WRKY1) 87 4e-18
At4g12020 putative disease resistance protein 79 1e-15
At2g44745 WRKY transcription factor 12 (WRKY12) 74 4e-14
At1g55600 putative protein 74 6e-14
At5g13080 WRKY-like protein 72 2e-13
At4g18170 DNA binding like protein 71 4e-13
At1g29860 putative protein 70 8e-13
At2g47260 putative WRKY-type DNA binding protein 69 1e-12
At5g46350 putative protein 69 1e-12
At3g01970 putative WRKY-like transcriptional regulator protein 69 1e-12
>At2g38470 putative WRKY-type DNA binding protein
Length = 512
Score = 110 bits (275), Expect = 4e-25
Identities = 57/127 (44%), Positives = 77/127 (59%), Gaps = 23/127 (18%)
Query: 60 NGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEV 119
NGR+Q K +DGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER+++G++
Sbjct: 169 NGREQRKG-------EDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSLEGQI 221
Query: 120 IETLYKGTHNHWKPTSSMKRNSSSEYLYSLL---------------PSETGSIDLQDQSF 164
E +YKG+HNH KP S+ + +SSS +S + P+ S Q SF
Sbjct: 222 TEIVYKGSHNHPKPQSTRRSSSSSSTFHSAVYNASLDHNRQASSDQPNSNNSFH-QSDSF 280
Query: 165 GSEQLDS 171
G +Q D+
Sbjct: 281 GMQQEDN 287
Score = 72.8 bits (177), Expect = 1e-13
Identities = 37/87 (42%), Positives = 51/87 (58%), Gaps = 6/87 (6%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 355 DDGYRWRKYGQKVVKGNPNPRSYYKCTTIGCPVRKHVERASHDMRAVITTYEGKHNHDVP 414
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQ 160
+ S Y + P ++ S+ ++
Sbjct: 415 AA-----RGSGYATNRAPQDSSSVPIR 436
>At5g07100 SPF1-like protein
Length = 309
Score = 97.1 bits (240), Expect = 5e-21
Identities = 47/81 (58%), Positives = 61/81 (75%), Gaps = 4/81 (4%)
Query: 66 KSSVRRMNH--DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERT-IDGEVIET 122
KS + N DDGYNW+KY +K KGSEN RSY+KCT+PNC KKKVE + + G++IE
Sbjct: 106 KSEIMSSNKTSDDGYNWRKYGQKQVKGSENPRSYFKCTYPNCLTKKKVETSLVKGQMIEI 165
Query: 123 LYKGTHNHWKPTSSMKRNSSS 143
+YKG+HNH KP S+ KR+SS+
Sbjct: 166 VYKGSHNHPKPQST-KRSSST 185
Score = 76.6 bits (187), Expect = 7e-15
Identities = 34/61 (55%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT+ CFV+K VER D + + T Y+G H H P
Sbjct: 234 DDGYRWRKYGQKVVKGNPNPRSYYKCTFTGCFVRKHVERAFQDPKSVITTYEGKHKHQIP 293
Query: 134 T 134
T
Sbjct: 294 T 294
>At1g13960 WRKY DNA-binding protein 4 (WRKY4)
Length = 514
Score = 95.9 bits (237), Expect = 1e-20
Identities = 48/109 (44%), Positives = 67/109 (61%), Gaps = 2/109 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPT 134
DDGYNW+KY +K KGSE RSYYKCT P C VKKKVER++DG+V E +YKG HNH P
Sbjct: 229 DDGYNWRKYGQKQVKGSEFPRSYYKCTNPGCPVKKKVERSLDGQVTEIIYKGQHNHEPPQ 288
Query: 135 SSMKRN--SSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSSVS 181
++ + N +++ S + + GS +L F + + + +VS
Sbjct: 289 NTKRGNKDNTANINGSSINNNRGSSELGASQFQTNSSNKTKREQHEAVS 337
Score = 75.1 bits (183), Expect = 2e-14
Identities = 42/105 (40%), Positives = 57/105 (54%), Gaps = 3/105 (2%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ RSYYKCT P C V+K VER D + + T Y+G HNH P
Sbjct: 409 DDGYRWRKYGQKVVKGNPYPRSYYKCTTPGCGVRKHVERAATDPKAVVTTYEGKHNHDLP 468
Query: 134 T--SSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPT 176
SS ++++ P +++ Q Q +L EE T
Sbjct: 469 AAKSSSHAAAAAQLRPDNRPGGLANLNQQQQQQPVARLRLKEEQT 513
>At2g37260 putative WRKY-type DNA binding protein
Length = 349
Score = 94.7 bits (234), Expect = 2e-20
Identities = 42/80 (52%), Positives = 58/80 (72%)
Query: 64 QNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETL 123
++++S + DGYNW+KY +K KGSE RSYYKCT P C VKKKVER+++G+V E +
Sbjct: 74 ESETSTGDRSSVDGYNWRKYGQKQVKGSECPRSYYKCTHPKCPVKKKVERSVEGQVSEIV 133
Query: 124 YKGTHNHWKPTSSMKRNSSS 143
Y+G HNH KP+ + R +SS
Sbjct: 134 YQGEHNHSKPSCPLPRRASS 153
Score = 64.7 bits (156), Expect = 3e-11
Identities = 32/89 (35%), Positives = 47/89 (51%), Gaps = 1/89 (1%)
Query: 43 EIDGVDNNRNEFQSDYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTW 102
E+D ++ S+ ++ SV + +DG+ W+KY +KV G+ RSYY+CT
Sbjct: 237 ELDDPSRSKRRKNEKQSSEAGVSQGSVESDSLEDGFRWRKYGQKVVGGNAYPRSYYRCTS 296
Query: 103 PNCFVKKKVERTIDG-EVIETLYKGTHNH 130
NC +K VER D T Y+G HNH
Sbjct: 297 ANCRARKHVERASDDPRAFITTYEGKHNH 325
>At2g03340 putative WRKY DNA-binding protein
Length = 513
Score = 94.4 bits (233), Expect = 3e-20
Identities = 47/109 (43%), Positives = 67/109 (61%), Gaps = 2/109 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPT 134
DDGYNW+KY +K KGS+ RSYYKCT P C VKKKVER++DG+V E +YKG HNH P
Sbjct: 250 DDGYNWRKYGQKQVKGSDFPRSYYKCTHPACPVKKKVERSLDGQVTEIIYKGQHNHELPQ 309
Query: 135 SSMKRNSS--SEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSSVS 181
N S S + + + S++ + + Q+ + E+ +++S S
Sbjct: 310 KRGNNNGSCKSSDIANQFQTSNSSLNKSKRDQETSQVTTTEQMSEASDS 358
Score = 73.6 bits (179), Expect = 6e-14
Identities = 33/62 (53%), Positives = 42/62 (67%), Gaps = 1/62 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ RSYYKCT P+C V+K VER D + + T Y+G HNH P
Sbjct: 415 DDGYRWRKYGQKVVKGNPYPRSYYKCTTPDCGVRKHVERAATDPKAVVTTYEGKHNHDVP 474
Query: 134 TS 135
+
Sbjct: 475 AA 476
>At4g26640 WRKY transcription factor 20 (WRKY20)
Length = 557
Score = 93.2 bits (230), Expect = 7e-20
Identities = 51/126 (40%), Positives = 69/126 (54%), Gaps = 8/126 (6%)
Query: 55 QSDYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERT 114
+ + ++ S + DDGYNW+KY +K KGSE RSYYKCT PNC VKK ER+
Sbjct: 191 EESIQTSQNDSRGSTPSILADDGYNWRKYGQKHVKGSEFPRSYYKCTHPNCEVKKLFERS 250
Query: 115 IDGEVIETLYKGTHNHWKPTSSMKRNS----SSEYLYSLLPSETGSIDLQDQSFGSEQLD 170
DG++ + +YKGTH+H KP +RNS + E PS TG ++ G L
Sbjct: 251 HDGQITDIIYKGTHDHPKPQPG-RRNSGGMAAQEERLDKYPSSTGR---DEKGSGVYNLS 306
Query: 171 SDEEPT 176
+ E T
Sbjct: 307 NPNEQT 312
Score = 77.8 bits (190), Expect = 3e-15
Identities = 41/86 (47%), Positives = 54/86 (62%), Gaps = 3/86 (3%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV +G+ N RSYYKCT C V+K VER + D + + T Y+G H+H P
Sbjct: 381 DDGYRWRKYGQKVVRGNPNPRSYYKCTAHGCPVRKHVERASHDPKAVITTYEGKHDHDVP 440
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDL 159
TS K +S+ E P ET +I L
Sbjct: 441 TS--KSSSNHEIQPRFRPDETDTISL 464
>At5g56270 transcription factor NtWRKY4-like
Length = 687
Score = 92.8 bits (229), Expect = 9e-20
Identities = 39/59 (66%), Positives = 47/59 (79%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKP 133
+DGYNW+KY +K+ KGSE RSYYKCT PNC VKKKVER+ +G + E +YKG HNH KP
Sbjct: 273 EDGYNWRKYGQKLVKGSEYPRSYYKCTNPNCQVKKKVERSREGHITEIIYKGAHNHLKP 331
Score = 75.9 bits (185), Expect = 1e-14
Identities = 38/69 (55%), Positives = 47/69 (68%), Gaps = 4/69 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT P C V+K VER + D + + T Y+G HNH P
Sbjct: 487 DDGYRWRKYGQKVVKGNPNPRSYYKCTAPGCTVRKHVERASHDLKSVITTYEGKHNHDVP 546
Query: 134 TSSMKRNSS 142
+ RNSS
Sbjct: 547 AA---RNSS 552
>At4g26440 putative protein
Length = 568
Score = 89.4 bits (220), Expect = 1e-18
Identities = 39/69 (56%), Positives = 48/69 (69%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPT 134
DDGYNW+KY +K+ KGSE RSYYKCT PNC KKKVER+ +G +IE +Y G H H KP
Sbjct: 178 DDGYNWRKYGQKLVKGSEYPRSYYKCTHPNCEAKKKVERSREGHIIEIIYTGDHIHSKPP 237
Query: 135 SSMKRNSSS 143
+ + S
Sbjct: 238 PNRRSGIGS 246
Score = 68.9 bits (167), Expect = 1e-12
Identities = 36/69 (52%), Positives = 42/69 (60%), Gaps = 4/69 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDG-EVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V K VER D + + T Y G H H P
Sbjct: 372 DDGYRWRKYGQKVVKGNPNPRSYYKCTANGCTVTKHVERASDDFKSVLTTYIGKHTHVVP 431
Query: 134 TSSMKRNSS 142
+ RNSS
Sbjct: 432 AA---RNSS 437
>At2g30250 putative WRKY-type DNA binding protein
Length = 393
Score = 88.6 bits (218), Expect = 2e-18
Identities = 51/122 (41%), Positives = 68/122 (54%), Gaps = 20/122 (16%)
Query: 73 NHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWK 132
N +DGY W+KY +K K SEN RSY+KCT+P+C KK VE DG++ E +YKG HNH K
Sbjct: 164 NSNDGYGWRKYGQKQVKKSENPRSYFKCTYPDCVSKKIVETASDGQITEIIYKGGHNHPK 223
Query: 133 P-------TSSMKRNSSSEYLYSLL--------PSETGSI-----DLQDQSFGSEQLDSD 172
P SS+ + + L++ SE SI DL+ +SF SE + D
Sbjct: 224 PEFTKRPSQSSLPSSVNGRRLFNPASVVSEPHDQSENSSISFDYSDLEQKSFKSEYGEID 283
Query: 173 EE 174
EE
Sbjct: 284 EE 285
Score = 73.6 bits (179), Expect = 6e-14
Identities = 33/61 (54%), Positives = 43/61 (70%), Gaps = 1/61 (1%)
Query: 76 DGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERT-IDGEVIETLYKGTHNHWKPT 134
DG+ W+KY +KV KG+ N RSYYKCT+ C VKK+VER+ D + T Y+G HNH PT
Sbjct: 329 DGFRWRKYGQKVVKGNTNPRSYYKCTFQGCGVKKQVERSAADERAVLTTYEGRHNHDIPT 388
Query: 135 S 135
+
Sbjct: 389 A 389
>At3g01080 WRKY transcription factor 58 (WRKY58)
Length = 423
Score = 88.2 bits (217), Expect = 2e-18
Identities = 44/96 (45%), Positives = 63/96 (64%), Gaps = 4/96 (4%)
Query: 41 AIEIDGVDNN-RNEFQS--DYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSY 97
++++ VD RN + + + +N R N +V + DDGYNW+KY +K KG E RSY
Sbjct: 131 SLDVSQVDQRARNHYNNPGNNNNNRSYNVVNVDKPA-DDGYNWRKYGQKPIKGCEYPRSY 189
Query: 98 YKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKP 133
YKCT NC VKKKVER+ DG++ + +YKG H+H +P
Sbjct: 190 YKCTHVNCPVKKKVERSSDGQITQIIYKGQHDHERP 225
Score = 79.0 bits (193), Expect = 1e-15
Identities = 38/73 (52%), Positives = 49/73 (67%), Gaps = 3/73 (4%)
Query: 64 QNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIET 122
Q KS V + DDGY W+KY +KV KG+ + RSYYKCT PNC V+K VER + D + + T
Sbjct: 297 QTKSEVDLL--DDGYRWRKYGQKVVKGNPHPRSYYKCTTPNCTVRKHVERASTDAKAVIT 354
Query: 123 LYKGTHNHWKPTS 135
Y+G HNH P +
Sbjct: 355 TYEGKHNHDVPAA 367
>At2g04880 WRKY transcription factor 1 splice variant 1 (WRKY1)
Length = 487
Score = 87.4 bits (215), Expect = 4e-18
Identities = 36/74 (48%), Positives = 53/74 (70%)
Query: 60 NGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEV 119
+G + N +R +DGYNW+KY +K+ KG+E RSYY+CT PNC KK++ER+ G+V
Sbjct: 96 SGSEGNSPFIREKVMEDGYNWRKYGQKLVKGNEFVRSYYRCTHPNCKAKKQLERSAGGQV 155
Query: 120 IETLYKGTHNHWKP 133
++T+Y G H+H KP
Sbjct: 156 VDTVYFGEHDHPKP 169
Score = 66.6 bits (161), Expect = 7e-12
Identities = 36/100 (36%), Positives = 57/100 (57%), Gaps = 3/100 (3%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
+DGY W+KY +K KGS RSYY+C+ P C VKK VER+ D +++ T Y+G H+H P
Sbjct: 307 NDGYRWRKYGQKSVKGSPYPRSYYRCSSPGCPVKKHVERSSHDTKLLITTYEGKHDHDMP 366
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDE 173
+ +++ L S + + G + QS + + D+
Sbjct: 367 PGRVVTHNN--MLDSEVDDKEGDANKTPQSSTLQSITKDQ 404
>At4g12020 putative disease resistance protein
Length = 1895
Score = 79.0 bits (193), Expect = 1e-15
Identities = 36/72 (50%), Positives = 49/72 (68%), Gaps = 2/72 (2%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPT 134
+DGYNW+KY +K KGS+ SYYKCT+ C K+KVER++DG+V E +YK HNH P
Sbjct: 468 NDGYNWQKYGQKKVKGSKFPLSYYKCTYLGCPSKRKVERSLDGQVAEIVYKDRHNHEPPN 527
Query: 135 SSMKRNSSSEYL 146
++ S+ YL
Sbjct: 528 QG--KDGSTTYL 537
Score = 33.5 bits (75), Expect = 0.065
Identities = 13/23 (56%), Positives = 16/23 (69%)
Query: 73 NHDDGYNWKKYEEKVAKGSENQR 95
N DDGY W+KY +KV KG+ R
Sbjct: 639 NLDDGYRWRKYGQKVVKGNPYPR 661
>At2g44745 WRKY transcription factor 12 (WRKY12)
Length = 218
Score = 73.9 bits (180), Expect = 4e-14
Identities = 44/119 (36%), Positives = 64/119 (52%), Gaps = 19/119 (15%)
Query: 31 IGTLRVEDDVAIEIDGVDNNRNE---FQSDYSNGRQQNKSSVRRMNH------------- 74
+G+ V DD G +N+ + ++S+ +G +NK +RR
Sbjct: 83 LGSKVVNDDQENFGGGTNNDAHSNSWWRSNSGSGDMKNKVKIRRKLREPRFCFQTKSDVD 142
Query: 75 --DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNH 130
DDGY W+KY +KV K S + RSYY+CT NC VKK+VER + D ++ T Y+G HNH
Sbjct: 143 VLDDGYKWRKYGQKVVKNSLHPRSYYRCTHNNCRVKKRVERLSEDCRMVITTYEGRHNH 201
>At1g55600 putative protein
Length = 485
Score = 73.6 bits (179), Expect = 6e-14
Identities = 35/71 (49%), Positives = 45/71 (63%), Gaps = 1/71 (1%)
Query: 73 NHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDG-EVIETLYKGTHNHW 131
N +DGY W+KY +KV KG+ N RSY+KCT C VKK VER D +++ T Y G HNH
Sbjct: 305 NPNDGYRWRKYGQKVVKGNPNPRSYFKCTNIECRVKKHVERGADNIKLVVTTYDGIHNHP 364
Query: 132 KPTSSMKRNSS 142
P + +SS
Sbjct: 365 SPPARRSNSSS 375
>At5g13080 WRKY-like protein
Length = 145
Score = 72.0 bits (175), Expect = 2e-13
Identities = 34/77 (44%), Positives = 50/77 (64%), Gaps = 3/77 (3%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNH--W 131
DDGY W+KY +K K ++ RSYY+CT+ C VKK+V+R T+D EV+ T Y+G H+H
Sbjct: 67 DDGYRWRKYGQKAVKNNKFPRSYYRCTYGGCNVKKQVQRLTVDQEVVVTTYEGVHSHPIE 126
Query: 132 KPTSSMKRNSSSEYLYS 148
K T + + + +YS
Sbjct: 127 KSTENFEHILTQMQIYS 143
>At4g18170 DNA binding like protein
Length = 318
Score = 70.9 bits (172), Expect = 4e-13
Identities = 34/75 (45%), Positives = 50/75 (66%), Gaps = 2/75 (2%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
+DGY W+KY +K K S RSYY+CT C VKK+VER+ D V+ T Y+G HNH P
Sbjct: 172 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCNVKKRVERSFQDPTVVITTYEGQHNHPIP 231
Query: 134 TSSMKRNSSSEYLYS 148
T +++ +S++ ++S
Sbjct: 232 T-NLRGSSAAAAMFS 245
>At1g29860 putative protein
Length = 282
Score = 69.7 bits (169), Expect = 8e-13
Identities = 33/73 (45%), Positives = 48/73 (65%), Gaps = 2/73 (2%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
+DGY W+KY +K K S RSYY+CT C VKK+VER+ D ++ T Y+G HNH P
Sbjct: 136 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCNVKKRVERSFQDPSIVITTYEGKHNHPIP 195
Query: 134 TSSMKRNSSSEYL 146
S+++ ++E+L
Sbjct: 196 -STLRGTVAAEHL 207
>At2g47260 putative WRKY-type DNA binding protein
Length = 337
Score = 69.3 bits (168), Expect = 1e-12
Identities = 34/95 (35%), Positives = 54/95 (56%), Gaps = 7/95 (7%)
Query: 49 NNRNEFQSDYSNGRQQNKSSVRRMNH------DDGYNWKKYEEKVAKGSENQRSYYKCTW 102
+ + + ++ +N ++Q ++ V M +DGY W+KY +K K S RSYY+CT
Sbjct: 142 HTKKQLKAKKNNQKRQREARVAFMTKSEVDHLEDGYRWRKYGQKAVKNSPFPRSYYRCTT 201
Query: 103 PNCFVKKKVERTI-DGEVIETLYKGTHNHWKPTSS 136
+C VKK+VER+ D + T Y+G H H P +S
Sbjct: 202 ASCNVKKRVERSFRDPSTVVTTYEGQHTHISPLTS 236
>At5g46350 putative protein
Length = 326
Score = 68.9 bits (167), Expect = 1e-12
Identities = 39/100 (39%), Positives = 55/100 (55%), Gaps = 5/100 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTI-DGEVIETLYKGTHNHWKP 133
+DGY W+KY +K K S RSYY+CT C VKK+VER+ D V+ T Y+ HNH P
Sbjct: 183 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCNVKKRVERSYQDPTVVITTYESQHNHPIP 242
Query: 134 ----TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQL 169
T+ ++S+Y S P + I +SF ++ L
Sbjct: 243 TNRRTAMFSGTTASDYNPSSSPIFSDLIINTPRSFSNDDL 282
>At3g01970 putative WRKY-like transcriptional regulator protein
Length = 147
Score = 68.9 bits (167), Expect = 1e-12
Identities = 38/100 (38%), Positives = 48/100 (48%), Gaps = 12/100 (12%)
Query: 43 EIDGVDNNRNEFQSDYSNGRQQNKSSVRRMNH-----------DDGYNWKKYEEKVAKGS 91
E GVDN+ S R + K R + DDGY W+KY +K K +
Sbjct: 22 EFHGVDNSAQPTTSSEEKPRSKKKKKEREARYAFQTRSQVDILDDGYRWRKYGQKAVKNN 81
Query: 92 ENQRSYYKCTWPNCFVKKKVERTIDGE-VIETLYKGTHNH 130
RSYYKCT C VKK+V+R E V+ T Y+G H H
Sbjct: 82 PFPRSYYKCTEEGCRVKKQVQRQWGDEGVVVTTYQGVHTH 121
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,212,625
Number of Sequences: 26719
Number of extensions: 179286
Number of successful extensions: 606
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 81
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 468
Number of HSP's gapped (non-prelim): 117
length of query: 182
length of database: 11,318,596
effective HSP length: 93
effective length of query: 89
effective length of database: 8,833,729
effective search space: 786201881
effective search space used: 786201881
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC138012.5


