

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137995.4 - phase: 0
(317 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
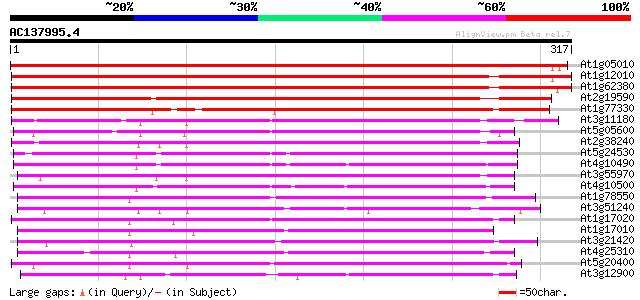
Score E
Sequences producing significant alignments: (bits) Value
At1g05010 1-aminocyclopropane-1-carboxylate oxidase 496 e-141
At1g12010 putative amino-cyclopropane-carboxylic acid oxidase (A... 472 e-133
At1g62380 Unknown protein (F24O1.10) 469 e-133
At2g19590 1-aminocyclopropane-1-carboxylate oxidase 304 5e-83
At1g77330 unknown protein 291 3e-79
At3g11180 putative leucoanthocyanidin dioxygenase 193 1e-49
At5g05600 leucoanthocyanidin dioxygenase-like protein 189 2e-48
At2g38240 putative anthocyanidin synthase 187 5e-48
At5g24530 flavanone 3-hydroxylase-like protein 183 9e-47
At4g10490 putative flavanone 3-beta-hydroxylase 180 8e-46
At3g55970 leucoanthocyanidin dioxygenase -like protein 171 6e-43
At4g10500 putative Fe(II)/ascorbate oxidase 169 1e-42
At1g78550 unknown protein 169 2e-42
At3g51240 flavanone 3-hydroxylase (FH3) 164 8e-41
At1g17020 SRG1-like protein 160 6e-40
At1g17010 SRG1-like protein 159 1e-39
At3g21420 unknown protein 157 9e-39
At4g25310 SRG1-like protein 156 1e-38
At5g20400 ethylene-forming-enzyme-like dioxygenase-like protein 155 2e-38
At3g12900 hypothetical protein 153 1e-37
>At1g05010 1-aminocyclopropane-1-carboxylate oxidase
Length = 323
Score = 496 bits (1277), Expect = e-141
Identities = 239/321 (74%), Positives = 287/321 (88%), Gaps = 6/321 (1%)
Query: 1 MMNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 60
M +FPII+LEKLNG ER TMEKIKDACENWGFFE VNHGI +L+D +E++TKEHY+KC
Sbjct: 1 MESFPIINLEKLNGEERAITMEKIKDACENWGFFECVNHGISLELLDKVEKMTKEHYKKC 60
Query: 61 MEHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLR 120
ME RFKE I ++GLD +++EV D+DWESTF+++HLP SNIS++PDL D+YR +MK+F+ +
Sbjct: 61 MEERFKESIKNRGLDSLRSEVNDVDWESTFYLKHLPVSNISDVPDLDDDYRTLMKDFAGK 120
Query: 121 LEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDA 180
+EKL+EELLDLLCENLGLEKGYLKK FYGS+ PTFGTKV+NYP CPNP+LVKGLRAHTDA
Sbjct: 121 IEKLSEELLDLLCENLGLEKGYLKKVFYGSKRPTFGTKVSNYPPCPNPDLVKGLRAHTDA 180
Query: 181 GGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQT 240
GGIILLFQDDKVSGLQLLKDG+WVDVPP++HSIVVNLGDQLEVITNGKYKSVEHRV++QT
Sbjct: 181 GGIILLFQDDKVSGLQLLKDGEWVDVPPVKHSIVVNLGDQLEVITNGKYKSVEHRVLSQT 240
Query: 241 NGT-RMSIASFYNPGSDAVIYPAPELLEKQTE-EKHNVYPKFVFEEYMKIYAALKFHAKE 298
+G RMSIASFYNPGSD+VI+PAPEL+ K+ E EK YP+FVFE+YMK+Y+A+KF AKE
Sbjct: 241 DGEGRMSIASFYNPGSDSVIFPAPELIGKEAEKEKKENYPRFVFEDYMKLYSAVKFQAKE 300
Query: 299 PRFEALK--ESNV--NLGPIA 315
PRFEA+K E+ V N+GP+A
Sbjct: 301 PRFEAMKAMETTVANNVGPLA 321
>At1g12010 putative amino-cyclopropane-carboxylic acid oxidase (ACC
oxidase)
Length = 320
Score = 472 bits (1215), Expect = e-133
Identities = 222/318 (69%), Positives = 268/318 (83%), Gaps = 7/318 (2%)
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
+ FP+I L KLNG ER TM I DAC+NWGFFELVNHG+P+DLMD +ER+TKEHY+K M
Sbjct: 5 IKFPVIDLSKLNGEERDQTMALIDDACQNWGFFELVNHGLPYDLMDNIERMTKEHYKKHM 64
Query: 62 EHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLRL 121
E +FKE++ SKGLD ++TEV+D+DWESTF++ HLP+SN+ +IPD+S+EYR MK+F RL
Sbjct: 65 EQKFKEMLRSKGLDTLETEVEDVDWESTFYLHHLPQSNLYDIPDMSNEYRLAMKDFGKRL 124
Query: 122 EKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDAG 181
E LAEELLDLLCENLGLEKGYLKK F+G+ GPTF TK++NYP CP PE++KGLRAHTDAG
Sbjct: 125 EILAEELLDLLCENLGLEKGYLKKVFHGTTGPTFATKLSNYPPCPKPEMIKGLRAHTDAG 184
Query: 182 GIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTN 241
G+ILLFQDDKVSGLQLLKDG WVDVPP++HSIV+NLGDQLEVITNGKYKSV HRV+ Q
Sbjct: 185 GLILLFQDDKVSGLQLLKDGDWVDVPPLKHSIVINLGDQLEVITNGKYKSVMHRVMTQKE 244
Query: 242 GTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPRF 301
G RMSIASFYNPGSDA I PA L++K ++ YP FVF++YMK+YA LKF AKEPRF
Sbjct: 245 GNRMSIASFYNPGSDAEISPATSLVDKDSK-----YPSFVFDDYMKLYAGLKFQAKEPRF 299
Query: 302 EALK--ESNVNLGPIAIV 317
EA+K E+ +L P+A+V
Sbjct: 300 EAMKNAEAAADLNPVAVV 317
>At1g62380 Unknown protein (F24O1.10)
Length = 320
Score = 469 bits (1207), Expect = e-133
Identities = 217/318 (68%), Positives = 263/318 (82%), Gaps = 7/318 (2%)
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
M FP++ L KLNG ER TM I +ACENWGFFE+VNHG+PHDLMD +E++TK+HY+ C
Sbjct: 5 MKFPVVDLSKLNGEERDQTMALINEACENWGFFEIVNHGLPHDLMDKIEKMTKDHYKTCQ 64
Query: 62 EHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLRL 121
E +F +++ SKGLD ++TEV+D+DWESTF+VRHLP+SN+++I D+SDEYR MK+F RL
Sbjct: 65 EQKFNDMLKSKGLDNLETEVEDVDWESTFYVRHLPQSNLNDISDVSDEYRTAMKDFGKRL 124
Query: 122 EKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDAG 181
E LAE+LLDLLCENLGLEKGYLKK F+G++GPTFGTKV+NYP CP PE++KGLRAHTDAG
Sbjct: 125 ENLAEDLLDLLCENLGLEKGYLKKVFHGTKGPTFGTKVSNYPPCPKPEMIKGLRAHTDAG 184
Query: 182 GIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRVIAQTN 241
GIILLFQDDKVSGLQLLKDG W+DVPP+ HSIV+NLGDQLEVITNGKYKSV HRV+ Q
Sbjct: 185 GIILLFQDDKVSGLQLLKDGDWIDVPPLNHSIVINLGDQLEVITNGKYKSVLHRVVTQQE 244
Query: 242 GTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHAKEPRF 301
G RMS+ASFYNPGSDA I PA L+EK +E YP FVF++YMK+YA +KF KEPRF
Sbjct: 245 GNRMSVASFYNPGSDAEISPATSLVEKDSE-----YPSFVFDDYMKLYAGVKFQPKEPRF 299
Query: 302 EALKESN--VNLGPIAIV 317
A+K ++ L P A V
Sbjct: 300 AAMKNASAVTELNPTAAV 317
>At2g19590 1-aminocyclopropane-1-carboxylate oxidase
Length = 310
Score = 304 bits (778), Expect = 5e-83
Identities = 147/307 (47%), Positives = 208/307 (66%), Gaps = 14/307 (4%)
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
M P+I +L+G +R TM + AC+ WGFF + NHGI +LM+ ++++ HY + +
Sbjct: 9 MEIPVIDFAELDGEKRSKTMSLLDHACDKWGFFMVDNHGIDKELMEKVKKMINSHYEEHL 68
Query: 62 EHRFKEVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSLRL 121
+ +F + K L +T D DWES+F + H P SNI +IP++S+E K M E+ +L
Sbjct: 69 KEKFYQSEMVKALSEGKTS--DADWESSFFISHKPTSNICQIPNISEELSKTMDEYVCQL 126
Query: 122 EKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRAHTDAG 181
K AE L L+CENLGL++ + AF G +GP FGTKVA YP+CP PEL++GLR HTDAG
Sbjct: 127 HKFAERLSKLMCENLGLDQEDIMNAFSGPKGPAFGTKVAKYPECPRPELMRGLREHTDAG 186
Query: 182 GIILLFQDDKVSGLQLLKDGQWVDVPPMR-HSIVVNLGDQLEVITNGKYKSVEHRVIAQT 240
GIILL QDD+V GL+ KDG+WV +PP + ++I VN GDQLE+++NG+YKSV HRV+
Sbjct: 187 GIILLLQDDQVPGLEFFKDGKWVPIPPSKNNTIFVNTGDQLEILSNGRYKSVVHRVMTVK 246
Query: 241 NGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPK-FVFEEYMKIYAALKFHAKEP 299
+G+R+SIA+FYNP DA+I PAP+LL YP + F++Y+K+Y+ KF K P
Sbjct: 247 HGSRLSIATFYNPAGDAIISPAPKLL----------YPSGYRFQDYLKLYSTTKFGDKGP 296
Query: 300 RFEALKE 306
R E +K+
Sbjct: 297 RLETMKK 303
>At1g77330 unknown protein
Length = 307
Score = 291 bits (745), Expect = 3e-79
Identities = 154/314 (49%), Positives = 213/314 (67%), Gaps = 19/314 (6%)
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
M P+I KLNG ER+ T+ +I ACE WGFF+LVNHGIP +L++ +++L+ + Y+
Sbjct: 1 MAIPVIDFSKLNGEEREKTLSEIARACEEWGFFQLVNHGIPLELLNKVKKLSSDCYKTER 60
Query: 62 EHRFKEVISSKGL-DVVQT----EVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKE 116
E FK K L ++VQ +++++DWE F L + N +E P ++ M E
Sbjct: 61 EEAFKTSNPVKLLNELVQKNSGEKLENVDWEDVFT---LLDHNQNEWPS---NIKETMGE 114
Query: 117 FSLRLEKLAEELLDLLCENLGLEKGYLKKAFY-----GSRGPTFGTKVANYPQCPNPELV 171
+ + KLA ++++++ ENLGL KGY+KKAF G FGTKV++YP CP+PELV
Sbjct: 115 YREEVRKLASKMMEVMDENLGLPKGYIKKAFNEGMEDGEETAFFGTKVSHYPPCPHPELV 174
Query: 172 KGLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKS 231
GLRAHTDAGG++LLFQDD+ GLQ+LKDG+W+DV P+ ++IV+N GDQ+EV++NG+YKS
Sbjct: 175 NGLRAHTDAGGVVLLFQDDEYDGLQVLKDGEWIDVQPLPNAIVINTGDQIEVLSNGRYKS 234
Query: 232 VEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAA 291
HRV+A+ G R SIASFYNP A I PA E+ +E+K YPKFVF +YM +YA
Sbjct: 235 AWHRVLAREEGNRRSIASFYNPSYKAAIGPAAVAEEEGSEKK---YPKFVFGDYMDVYAN 291
Query: 292 LKFHAKEPRFEALK 305
KF KEPRF A+K
Sbjct: 292 QKFMPKEPRFLAVK 305
>At3g11180 putative leucoanthocyanidin dioxygenase
Length = 400
Score = 193 bits (490), Expect = 1e-49
Identities = 112/318 (35%), Positives = 174/318 (54%), Gaps = 21/318 (6%)
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
+N PII L+ L +D ++I +AC WGFF+++NHG+ +LMD K + +
Sbjct: 93 INIPIIDLDSLFS-GNEDDKKRISEACREWGFFQVINHGVKPELMDAARETWKSFFNLPV 151
Query: 62 EHRFKEVISSK-------GLDVVQTEVKDMDWESTFHVRHLPES--NISEIPDLSDEYRK 112
E KEV S+ G + + +DW +++ LP + + ++ P L R+
Sbjct: 152 EA--KEVYSNSPRTYEGYGSRLGVEKGAILDWNDYYYLHFLPLALKDFNKWPSLPSNIRE 209
Query: 113 VMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVK 172
+ E+ L KL L+ +L NLGL L++AF G +V YP+CP PEL
Sbjct: 210 MNDEYGKELVKLGGRLMTILSSNLGLRAEQLQEAF-GGEDVGACLRVNYYPKCPQPELAL 268
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSV 232
GL H+D GG+ +L DD+V GLQ+ W+ V P+RH+ +VN+GDQ+++++N KYKSV
Sbjct: 269 GLSPHSDPGGMTILLPDDQVVGLQVRHGDTWITVNPLRHAFIVNIGDQIQILSNSKYKSV 328
Query: 233 EHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAAL 292
EHRVI + R+S+A FYNP SD I P +L+ T +YP F++Y L
Sbjct: 329 EHRVIVNSEKERVSLAFFYNPKSDIPIQPMQQLV---TSTMPPLYPPMTFDQY-----RL 380
Query: 293 KFHAKEPRFEALKESNVN 310
+ PR ++ ES+++
Sbjct: 381 FIRTQGPRGKSHVESHIS 398
>At5g05600 leucoanthocyanidin dioxygenase-like protein
Length = 371
Score = 189 bits (479), Expect = 2e-48
Identities = 109/297 (36%), Positives = 162/297 (53%), Gaps = 22/297 (7%)
Query: 3 NFPIISLEKL---NGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRK 59
N PII LE L G+ M +I +AC WGFF++VNHG+ +LMD +E +
Sbjct: 61 NIPIIDLEGLFSEEGLSDDVIMARISEACRGWGFFQVVNHGVKPELMDAARENWREFFH- 119
Query: 60 CMEHRFKEVISSK-------GLDVVQTEVKDMDWESTFHVRHLPE--SNISEIPDLSDEY 110
M KE S+ G + + +DW + + LP + ++ P
Sbjct: 120 -MPVNAKETYSNSPRTYEGYGSRLGVEKGASLDWSDYYFLHLLPHHLKDFNKWPSFPPTI 178
Query: 111 RKVMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPEL 170
R+V+ E+ L KL+ ++ +L NLGL++ ++AF G +V YP+CP PEL
Sbjct: 179 REVIDEYGEELVKLSGRIMRVLSTNLGLKEDKFQEAF-GGENIGACLRVNYYPKCPRPEL 237
Query: 171 VKGLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYK 230
GL H+D GG+ +L DD+V GLQ+ KD W+ V P H+ +VN+GDQ+++++N YK
Sbjct: 238 ALGLSPHSDPGGMTILLPDDQVFGLQVRKDDTWITVKPHPHAFIVNIGDQIQILSNSTYK 297
Query: 231 SVEHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHN--VYPKFVFEEY 285
SVEHRVI ++ R+S+A FYNP SD I P EL+ HN +YP F++Y
Sbjct: 298 SVEHRVIVNSDKERVSLAFFYNPKSDIPIQPLQELV-----STHNPPLYPPMTFDQY 349
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 187 bits (476), Expect = 5e-48
Identities = 101/294 (34%), Positives = 168/294 (56%), Gaps = 12/294 (4%)
Query: 2 MNFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
+ P++ + + G + + + ++ ACE WGFF++VNHG+ H LM+ + +E + +
Sbjct: 46 IEIPVLDMNDVWG--KPEGLRLVRSACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPL 103
Query: 62 EHRFKEVISS---KGLDVVQTEVKD--MDWESTFHVRHLPES--NISEIPDLSDEYRKVM 114
E + K S +G VKD +DW F + +LP S N S+ P + R+++
Sbjct: 104 EEKRKYANSPDTYEGYGSRLGVVKDAKLDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELI 163
Query: 115 KEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGL 174
+++ + KL E L + L E+LGL+ L +A G + YP+CP P+L GL
Sbjct: 164 EKYGEEVRKLCERLTETLSESLGLKPNKLMQALGGGDKVGASLRTNFYPKCPQPQLTLGL 223
Query: 175 RAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEH 234
+H+D GGI +L D+KV+GLQ+ + WV + + ++++VN+GDQL++++NG YKSVEH
Sbjct: 224 SSHSDPGGITILLPDEKVAGLQVRRGDGWVTIKSVPNALIVNIGDQLQILSNGIYKSVEH 283
Query: 235 RVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKI 288
+VI + R+S+A FYNP SD + P EL+ T + +Y F+EY +
Sbjct: 284 QVIVNSGMERVSLAFFYNPRSDIPVGPIEELV---TANRPALYKPIRFDEYRSL 334
>At5g24530 flavanone 3-hydroxylase-like protein
Length = 341
Score = 183 bits (465), Expect = 9e-47
Identities = 102/294 (34%), Positives = 160/294 (53%), Gaps = 17/294 (5%)
Query: 3 NFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCME 62
+FP+I L + +R +++I AC +GFF+++NHG+ ++D + + +E + ME
Sbjct: 37 DFPLIDL---SSTDRSFLIQQIHQACARFGFFQVINHGVNKQIIDEMVSVAREFFSMSME 93
Query: 63 HRFKEVIS--------SKGLDVVQTEVKDMDWESTFHVRHLP-ESNISEIPDLSDEYRKV 113
+ K S +V + EV + W + P ++E P ++++
Sbjct: 94 EKMKLYSDDPTKTTRLSTSFNVKKEEVNN--WRDYLRLHCYPIHKYVNEWPSNPPSFKEI 151
Query: 114 MKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKG 173
+ ++S + ++ ++ +L+ E+LGLEK Y+KK G +G V YP CP PEL G
Sbjct: 152 VSKYSREVREVGFKIEELISESLGLEKDYMKKVL-GEQGQHMA--VNYYPPCPEPELTYG 208
Query: 174 LRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
L AHTD + +L QD V GLQ+L DGQW V P + V+N+GDQL+ ++NG YKSV
Sbjct: 209 LPAHTDPNALTILLQDTTVCGLQILIDGQWFAVNPHPDAFVINIGDQLQALSNGVYKSVW 268
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMK 287
HR + T R+S+ASF P AV+ PA L E + +E VY F + EY K
Sbjct: 269 HRAVTNTENPRLSVASFLCPADCAVMSPAKPLWEAEDDETKPVYKDFTYAEYYK 322
>At4g10490 putative flavanone 3-beta-hydroxylase
Length = 348
Score = 180 bits (457), Expect = 8e-46
Identities = 104/294 (35%), Positives = 161/294 (54%), Gaps = 16/294 (5%)
Query: 3 NFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCME 62
+ P+I L L+G R D + + AC + GFF++ NHG+P + + + +E +R+
Sbjct: 41 SIPLIDLHDLHGPNRADIINQFAHACSSCGFFQIKNHGVPEETIKKMMNAAREFFRQSES 100
Query: 63 HRFKEVIS--------SKGLDVVQTEVKDMDWESTFHVRHLP-ESNISEIPDLSDEYRKV 113
R K + S +V + +V + W + P E I+E P +R+V
Sbjct: 101 ERVKHYSADTKKTTRLSTSFNVSKEKVSN--WRDFLRLHCYPIEDFINEWPSTPISFREV 158
Query: 114 MKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKG 173
E++ + L LL+ + E+LGL K + G G + YP+CP PEL G
Sbjct: 159 TAEYATSVRALVLTLLEAISESLGLAKDRVSNTI-GKHGQHMA--INYYPRCPQPELTYG 215
Query: 174 LRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
L H DA I +L QD+ VSGLQ+ KDG+W+ V P+ ++ +VNLGDQ++VI+N KYKSV
Sbjct: 216 LPGHKDANLITVLLQDE-VSGLQVFKDGKWIAVNPVPNTFIVNLGDQMQVISNEKYKSVL 274
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMK 287
HR + ++ R+SI +FY P DAVI PA EL+ ++ E+ +Y F + EY +
Sbjct: 275 HRAVVNSDMERISIPTFYCPSEDAVISPAQELINEE-EDSPAIYRNFTYAEYFE 327
>At3g55970 leucoanthocyanidin dioxygenase -like protein
Length = 363
Score = 171 bits (432), Expect = 6e-43
Identities = 96/292 (32%), Positives = 160/292 (53%), Gaps = 14/292 (4%)
Query: 5 PIISLEKLNGVE---RKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
PII L +L + + T+++I AC GFF++VNHG+ LMD + +E + M
Sbjct: 53 PIIDLGRLYTDDLTLQAKTLDEISKACRELGFFQVVNHGMSPQLMDQAKATWREFFNLPM 112
Query: 62 EHRFKEVISSKGLDVVQTEV-----KDMDWESTFHVRHLPES--NISEIPDLSDEYRKVM 114
E + S K + + + +DW +++ + P S + ++ P L R+++
Sbjct: 113 ELKNMHANSPKTYEGYGSRLGVEKGAILDWSDYYYLHYQPSSLKDYTKWPSLPLHCREIL 172
Query: 115 KEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGL 174
+++ + KL E L+ +L +NLGL++ L+ AF G +V YP+CP PEL G+
Sbjct: 173 EDYCKEMVKLCENLMKILSKNLGLQEDRLQNAFGGKEESGGCLRVNYYPKCPQPELTLGI 232
Query: 175 RAHTDAGGIILLFQDDKVSGLQLL-KDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
H+D GG+ +L D++V+ LQ+ D W+ V P H+ +VN+GDQ+++++N YKSVE
Sbjct: 233 SPHSDPGGLTILLPDEQVASLQVRGSDDAWITVEPAPHAFIVNMGDQIQMLSNSIYKSVE 292
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEY 285
HRVI R+S+A FYNP + I P EL+ T + +Y ++ Y
Sbjct: 293 HRVIVNPENERLSLAFFYNPKGNVPIEPLKELV---TVDSPALYSSTTYDRY 341
>At4g10500 putative Fe(II)/ascorbate oxidase
Length = 349
Score = 169 bits (429), Expect = 1e-42
Identities = 100/292 (34%), Positives = 156/292 (53%), Gaps = 17/292 (5%)
Query: 3 NFPIISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCME 62
+ P+I L L+G R ++++ AC +GFF++ NHG+P ++ ++ + +E + +
Sbjct: 43 SIPLIDLRDLHGPNRAVIVQQLASACSTYGFFQIKNHGVPDTTVNKMQTVAREFFHQPES 102
Query: 63 HRFKEVIS--------SKGLDVVQTEVKDMDWESTFHVRHLP-ESNISEIPDLSDEYRKV 113
R K + S +V +V ++W + P E I E P +R+V
Sbjct: 103 ERVKHYSADPTKTTRLSTSFNVGADKV--LNWRDFLRLHCFPIEDFIEEWPSSPISFREV 160
Query: 114 MKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKG 173
E++ + L LL+ + E+LGLE ++ G YP CP PEL G
Sbjct: 161 TAEYATSVRALVLRLLEAISESLGLESDHISNIL-GKHAQHMAFNY--YPPCPEPELTYG 217
Query: 174 LRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
L H D I +L QD +VSGLQ+ KD +WV V P+ ++ +VN+GDQ++VI+N KYKSV
Sbjct: 218 LPGHKDPTVITVLLQD-QVSGLQVFKDDKWVAVSPIPNTFIVNIGDQMQVISNDKYKSVL 276
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEY 285
HR + T R+SI +FY P +DAVI PA EL+ +Q + +Y + F EY
Sbjct: 277 HRAVVNTENERLSIPTFYFPSTDAVIGPAHELVNEQ--DSLAIYRTYPFVEY 326
>At1g78550 unknown protein
Length = 356
Score = 169 bits (428), Expect = 2e-42
Identities = 96/301 (31%), Positives = 166/301 (54%), Gaps = 14/301 (4%)
Query: 5 PIISLEKLNGVERKDT-MEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEH 63
P+I + +L V D+ ++K+ AC++WGFF+LVNHGI ++ LE +E + M+
Sbjct: 54 PVIDMTRLCSVSAMDSELKKLDFACQDWGFFQLVNHGIDSSFLEKLETEVQEFFNLPMKE 113
Query: 64 RFK-----EVISSKGLDVVQTEVKDMDWESTFHVRHLP-ESNISEI-PDLSDEYRKVMKE 116
+ K G + +E + +DW F + P S S + L +R+ ++
Sbjct: 114 KQKLWQRSGEFEGFGQVNIVSENQKLDWGDMFILTTEPIRSRKSHLFSKLPPPFRETLET 173
Query: 117 FSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRA 176
+S ++ +A+ L + L ++ ++ F K+ YP CP P+ V GL
Sbjct: 174 YSSEVKSIAKILFAKMASVLEIKHEEMEDLF---DDVWQSIKINYYPPCPQPDQVMGLTQ 230
Query: 177 HTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRV 236
H+DA G+ +L Q ++V GLQ+ KDG+WV V P+R ++VVN+G+ LE+ITNG+Y+S+EHR
Sbjct: 231 HSDAAGLTILLQVNQVEGLQIKKDGKWVVVKPLRDALVVNVGEILEIITNGRYRSIEHRA 290
Query: 237 IAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALKFHA 296
+ + R+S+A F++PG + +I PA L+++Q K ++ +EY + K +
Sbjct: 291 VVNSEKERLSVAMFHSPGKETIIRPAKSLVDRQ---KQCLFKSMSTQEYFDAFFTQKLNG 347
Query: 297 K 297
K
Sbjct: 348 K 348
>At3g51240 flavanone 3-hydroxylase (FH3)
Length = 358
Score = 164 bits (414), Expect = 8e-41
Identities = 111/314 (35%), Positives = 167/314 (52%), Gaps = 26/314 (8%)
Query: 5 PIISLEKLNGVERK--DTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCME 62
P+ISL ++ V+ K + +I +ACENWG F++V+HG+ +L+ + RL ++ + E
Sbjct: 39 PVISLAGIDDVDGKRGEICRQIVEACENWGIFQVVDHGVDTNLVADMTRLARDFFALPPE 98
Query: 63 HRFKEVISS--KGLDVVQTEVKD---MDWESTFHVRHLPESN--ISEIPDLSDEYRKVMK 115
+ + +S KG +V + ++ DW P N S PD + + KV +
Sbjct: 99 DKLRFDMSGGKKGGFIVSSHLQGEAVQDWREIVTYFSYPVRNRDYSRWPDKPEGWVKVTE 158
Query: 116 EFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLR 175
E+S RL LA +LL++L E +GLEK L A V YP+CP P+L GL+
Sbjct: 159 EYSERLMSLACKLLEVLSEAMGLEKESLTNACVDMDQKIV---VNYYPKCPQPDLTLGLK 215
Query: 176 AHTDAGGIILLFQDDKVSGLQLLKDG--QWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
HTD G I LL QD +V GLQ +D W+ V P+ + VVNLGD ++NG++K+ +
Sbjct: 216 RHTDPGTITLLLQD-QVGGLQATRDNGKTWITVQPVEGAFVVNLGDHGHFLSNGRFKNAD 274
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMK------ 287
H+ + +N +R+SIA+F NP DA +YP L+ + EK + F E K
Sbjct: 275 HQAVVNSNSSRLSIATFQNPAPDATVYP----LKVREGEKAILEEPITFAEMYKRKMGRD 330
Query: 288 -IYAALKFHAKEPR 300
A LK AKE R
Sbjct: 331 LELARLKKLAKEER 344
>At1g17020 SRG1-like protein
Length = 358
Score = 160 bits (406), Expect = 6e-40
Identities = 94/292 (32%), Positives = 159/292 (54%), Gaps = 12/292 (4%)
Query: 2 MNFPIISLEKLNGVERKDT-MEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKC 60
+ PII +++L D+ +EK+ AC+ WGFF+LVNHGI +D ++ ++ +
Sbjct: 51 IEIPIIDMKRLCSSTTMDSEVEKLDFACKEWGFFQLVNHGIDSSFLDKVKSEIQDFFNLP 110
Query: 61 MEHRFK-----EVISSKGLDVVQTEVKDMDWESTFH--VRHLPESNISEIPDLSDEYRKV 113
ME + K + I G V +E + +DW F V+ + P L +R
Sbjct: 111 MEEKKKFWQRPDEIEGFGQAFVVSEDQKLDWADLFFHTVQPVELRKPHLFPKLPLPFRDT 170
Query: 114 MKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKG 173
++ +S ++ +A+ L+ + L ++ L+K F ++ YP CP P+ V G
Sbjct: 171 LEMYSSEVQSVAKILIAKMARALEIKPEELEKLF-DDVDSVQSMRMNYYPPCPQPDQVIG 229
Query: 174 LRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
L H+D+ G+ +L Q + V GLQ+ KDG+WV V P+ ++ +VN+GD LE+ITNG Y+S+E
Sbjct: 230 LTPHSDSVGLTVLMQVNDVEGLQIKKDGKWVPVKPLPNAFIVNIGDVLEIITNGTYRSIE 289
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEY 285
HR + + R+SIA+F+N G + PA L+E+Q + + + +EY
Sbjct: 290 HRGVVNSEKERLSIATFHNVGMYKEVGPAKSLVERQKVAR---FKRLTMKEY 338
>At1g17010 SRG1-like protein
Length = 361
Score = 159 bits (403), Expect = 1e-39
Identities = 93/277 (33%), Positives = 154/277 (55%), Gaps = 10/277 (3%)
Query: 5 PIISLEKLNGVERKDT-MEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEH 63
PII + +L D+ +EK+ AC+ +GFF+LVNHGI +D ++ ++ + ME
Sbjct: 55 PIIDMNRLCSSTAVDSEVEKLDFACKEYGFFQLVNHGIDPSFLDKIKSEIQDFFNLPMEE 114
Query: 64 RFK-----EVISSKGLDVVQTEVKDMDWESTFHVRHLPESNISE--IPDLSDEYRKVMKE 116
+ K V+ G V +E + +DW F + P P L +R +
Sbjct: 115 KKKLWQTPAVMEGFGQAFVVSEDQKLDWADLFFLIMQPVQLRKRHLFPKLPLPFRDTLDM 174
Query: 117 FSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRA 176
+S R++ +A+ LL + + L ++ +++ F + ++ YP CP P LV GL
Sbjct: 175 YSTRVKSIAKILLAKMAKALQIKPEEVEEIFGDDMMQSM--RMNYYPPCPQPNLVTGLIP 232
Query: 177 HTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRV 236
H+DA G+ +L Q ++V GLQ+ K+G+W V P++++ +VN+GD LE+ITNG Y+S+EHR
Sbjct: 233 HSDAVGLTILLQVNEVDGLQIKKNGKWFFVKPLQNAFIVNVGDVLEIITNGTYRSIEHRA 292
Query: 237 IAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEK 273
+ R+SIA+F+N G D I PA L+++Q K
Sbjct: 293 MVNLEKERLSIATFHNTGMDKEIGPARSLVQRQEAAK 329
>At3g21420 unknown protein
Length = 364
Score = 157 bits (396), Expect = 9e-39
Identities = 91/305 (29%), Positives = 164/305 (52%), Gaps = 16/305 (5%)
Query: 5 PIISLEKLNGVERKD---TMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCM 61
P+I L KL+ + D + K+ ACE+WGFF+++NHGI ++++ +E + E + +
Sbjct: 56 PVIDLSKLSKPDNDDFFFEILKLSQACEDWGFFQVINHGIEVEVVEDIEEVASEFFDMPL 115
Query: 62 EHRFKE-----VISSKGLDVVQTEVKDMDWESTFHVR-HLPESNISEI-PDLSDEYRKVM 114
E + K + G + +E + +DW + F + H P+ ++ P + + +
Sbjct: 116 EEKKKYPMEPGTVQGYGQAFIFSEDQKLDWCNMFALGVHPPQIRNPKLWPSKPARFSESL 175
Query: 115 KEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGL 174
+ +S + +L + LL + +LGL++ ++ F + ++ YP C +P+LV GL
Sbjct: 176 EGYSKEIRELCKRLLKYIAISLGLKEERFEEMFGEA---VQAVRMNYYPPCSSPDLVLGL 232
Query: 175 RAHTDAGGIILLFQD-DKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVE 233
H+D + +L Q + GLQ+LKD WV V P+ +++V+N+GD +EV++NGKYKSVE
Sbjct: 233 SPHSDGSALTVLQQSKNSCVGLQILKDNTWVPVKPLPNALVINIGDTIEVLSNGKYKSVE 292
Query: 234 HRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIYAALK 293
HR + R++I +FY P + I P EL++ +T Y + +Y Y + K
Sbjct: 293 HRAVTNREKERLTIVTFYAPNYEVEIEPMSELVDDETNPCK--YRSYNHGDYSYHYVSNK 350
Query: 294 FHAKE 298
K+
Sbjct: 351 LQGKK 355
>At4g25310 SRG1-like protein
Length = 353
Score = 156 bits (395), Expect = 1e-38
Identities = 96/289 (33%), Positives = 154/289 (53%), Gaps = 16/289 (5%)
Query: 5 PIISLEKLNGVERKDT-MEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEH 63
PII + L+ D+ ++K+ AC+ WGFF+LVNHG+ +D + ++ + ME
Sbjct: 53 PIIDMSLLSSSTSMDSEIDKLDFACKEWGFFQLVNHGMD---LDKFKSDIQDFFNLPMEE 109
Query: 64 RFK-----EVISSKGLDVVQTEVKDMDWESTFHV--RHLPESNISEIPDLSDEYRKVMKE 116
+ K I G V +E + +DW F + + +P P L +R +
Sbjct: 110 KKKLWQQPGDIEGFGQAFVFSEEQKLDWADVFFLTMQPVPLRKPHLFPKLPLPFRDTLDT 169
Query: 117 FSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVKGLRA 176
+S L+ +A+ L L L ++ ++K F G ++ YP CP P+ GL
Sbjct: 170 YSAELKSIAKVLFAKLASALKIKPEEMEKLFDDELGQRI--RMNYYPPCPEPDKAIGLTP 227
Query: 177 HTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHRV 236
H+DA G+ +L Q ++V GLQ+ KDG+WV V P+ +++VVN+GD LE+ITNG Y+S+EHR
Sbjct: 228 HSDATGLTILLQVNEVEGLQIKKDGKWVSVKPLPNALVVNVGDILEIITNGTYRSIEHRG 287
Query: 237 IAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEY 285
+ + R+S+ASF+N G I P L+E+ K ++ EEY
Sbjct: 288 VVNSEKERLSVASFHNTGFGKEIGPMRSLVER---HKGALFKTLTTEEY 333
>At5g20400 ethylene-forming-enzyme-like dioxygenase-like protein
Length = 348
Score = 155 bits (393), Expect = 2e-38
Identities = 89/297 (29%), Positives = 151/297 (49%), Gaps = 12/297 (4%)
Query: 2 MNFPIISLEKL--NGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRK 59
M+ P I L L + + + K+ A WG +++NHGI +D + +LTKE +
Sbjct: 43 MDIPAIDLNLLLSSSEAGQQELSKLHSALSTWGVVQVMNHGITKAFLDKIYKLTKEFFAL 102
Query: 60 CMEHRFK-----EVISSKGLDVVQTEVKDMDWESTFHVRHLPESN--ISEIPDLSDEYRK 112
E + K + I G D++ + + +DW ++ PE ++ P++ +R+
Sbjct: 103 PTEEKQKCAREIDSIQGYGNDMILWDDQVLDWIDRLYITTYPEDQRQLNFWPEVPLGFRE 162
Query: 113 VMKEFSLRLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVANYPQCPNPELVK 172
+ E++++ + E+ + +L LE+ + T T+ YP CP+P+ V
Sbjct: 163 TLHEYTMKQRIVIEQFFKAMARSLELEENSFLDMY--GESATLDTRFNMYPPCPSPDKVI 220
Query: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSV 232
G++ H D I LL D V GLQ KDG+W P + +I++N+GDQ+E+++NG YKS
Sbjct: 221 GVKPHADGSAITLLLPDKDVGGLQFQKDGKWYKAPIVPDTILINVGDQMEIMSNGIYKSP 280
Query: 233 EHRVIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYMKIY 289
HRV+ R+S+A+F PG+D I P EL+ + + K+V E Y K Y
Sbjct: 281 VHRVVTNREKERISVATFCIPGADKEIQPVNELVSEARPRLYKTVKKYV-ELYFKYY 336
>At3g12900 hypothetical protein
Length = 357
Score = 153 bits (387), Expect = 1e-37
Identities = 92/291 (31%), Positives = 160/291 (54%), Gaps = 23/291 (7%)
Query: 7 ISLEKLNGVERKDTMEKIKDACENWGFFELVNHGIPHDLMDTLERLTKEHYRKCMEHR-- 64
I L L+G + K+ ++I +A E GFF++VNHG+ +L++ L+ E + + E +
Sbjct: 59 IDLSNLDGPQHKEVAKQIVEAAETLGFFQVVNHGVSVELLELLKSSAHEFFAQAPEEKSM 118
Query: 65 -FKEVISSK----GLDVVQTEVKDMDWESTFHVRHLPESNISEIPDLSDEYRKVMKEFSL 119
KEV SK G V + K ++W+ +V L ++ + R+V EF
Sbjct: 119 YLKEVSPSKLVKYGTSFVPDKEKAIEWKD--YVSMLYTNDSEALQHWPQPCREVALEFLN 176
Query: 120 RLEKLAEELLDLLCENLGLEKGYLKKAFYGSRGPTFGTKVAN---YPQCPNPELVKGLRA 176
++ + ++++L EN+G+ K GTK+ N YP CP+PEL G+
Sbjct: 177 SSMEMVKNVVNILMENVGVTLEEEKM------NGLMGTKMVNMNYYPTCPSPELTVGVGR 230
Query: 177 HTDAGGIILLFQDDKVSGLQL-LKDGQWVDVPPMRHSIVVNLGDQLEVITNGKYKSVEHR 235
H+D G + +L QD + GL + L +G+W ++PP+ ++V+N+GD L++++NGKYKS EHR
Sbjct: 231 HSDMGMLTVLLQDG-IGGLYVKLDNGEWAEIPPVHGALVINIGDTLQILSNGKYKSAEHR 289
Query: 236 VIAQTNGTRMSIASFYNPGSDAVIYPAPELLEKQTEEKHNVYPKFVFEEYM 286
V G+R+S+ F P + P PE++++ + Y +F+F++YM
Sbjct: 290 VRTTNIGSRVSVPIFTAPNPSQKVGPLPEVVKRDGVAR---YKEFLFQDYM 337
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,471,097
Number of Sequences: 26719
Number of extensions: 330897
Number of successful extensions: 1210
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 915
Number of HSP's gapped (non-prelim): 140
length of query: 317
length of database: 11,318,596
effective HSP length: 99
effective length of query: 218
effective length of database: 8,673,415
effective search space: 1890804470
effective search space used: 1890804470
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137995.4


