

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137995.11 + phase: 0
(1094 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
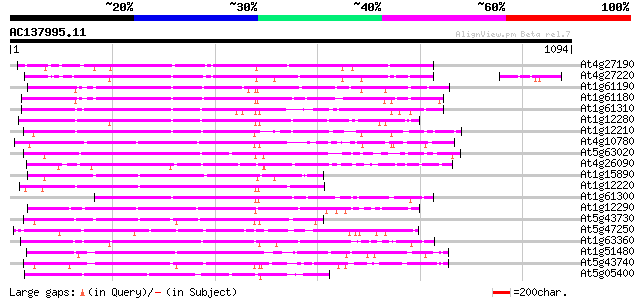
Score E
Sequences producing significant alignments: (bits) Value
At4g27190 putative protein 219 9e-57
At4g27220 putative protein 208 2e-53
At1g61190 206 6e-53
At1g61180 204 2e-52
At1g61310 similar to disease resistance protein 202 6e-52
At1g12280 hypothetical protein 202 6e-52
At1g12210 NBS/LRR disease resistance protein 192 7e-49
At4g10780 putative disease resistance protein 192 9e-49
At5g63020 NBS/LRR disease resistance like protein 191 3e-48
At4g26090 disease resistance protein RPS2 190 4e-48
At1g15890 hypothetical protein 185 1e-46
At1g12220 NBS/LRR disease resistance protein, putative 182 9e-46
At1g61300 similar to disease resistance protein 180 4e-45
At1g12290 hypothetical protein 178 1e-44
At5g43730 disease resistance protein 170 4e-42
At5g47250 NBS/LRR disease resistance protein 170 5e-42
At1g63360 disease resistance protein, putative 168 2e-41
At1g51480 hypothetical protein 167 3e-41
At5g43740 disease resistance protein 167 4e-41
At5g05400 NBS/LRR disease resistance protein 165 1e-40
>At4g27190 putative protein
Length = 985
Score = 219 bits (557), Expect = 9e-57
Identities = 236/900 (26%), Positives = 397/900 (43%), Gaps = 118/900 (13%)
Query: 16 VCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVV------- 68
V G I +L Y F+ V + K +SN+ A+ +S++ R+T K + E +
Sbjct: 7 VIGEILRLMYESTFSR-VANAIKFKSNVKALNESLE-RLTELKGNMSEDHETLLTKDKPL 64
Query: 69 ----EKWLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYI 124
+W ++A + V ++ E+ SC R + RKL K +K+
Sbjct: 65 RLKLMRWQREA----EEVISKARLKLEERVSC-----GMSLRPRMSRKLVKILDEVKMLE 115
Query: 125 EEGRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMC--------ALKDDDVTMIGL 176
++G +++++ S + ER ++ + L + IG+
Sbjct: 116 KDGIEFVDMLSVES------TPERVEHVPGVSVVHQTMASNMLAKIRDGLTSEKAQKIGV 169
Query: 177 YGMGGCGKTMLAMEVGKR-----CGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQ-E 230
+GMGG GKT L + + F V+FV +S + +Q++IA L+ + Q E
Sbjct: 170 WGMGGVGKTTLVRTLNNKLREEGATQPFGLVIFVIVSKEFDPREVQKQIAERLDIDTQME 229
Query: 231 KDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCT 290
+ E ++R+ + L +E + L+ILDDVW+ +D D +GIP E +KG K+++TSR VC
Sbjct: 230 ESEEKLARRIYVGLMKERKFLLILDDVWKPIDLDLLGIPRTEENKGSKVILTSRFLEVCR 289
Query: 291 LMDCQKKIQLSTLTNDETWDLFQKQALISEGTWI---SIKNMAREISNECKGLPVATVAV 347
M +++ L ++ W+LF K A G + ++ +A+ +S EC GLP+A + V
Sbjct: 290 SMKTDLDVRVDCLLEEDAWELFCKNA----GDVVRSDHVRKIAKAVSQECGGLPLAIITV 345
Query: 348 ASSLKGKAEVE-WKVALDRLRSSKP--VNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLC 404
++++GK V+ W L +L S P +IE+ + P L+LSYD L+ ++AK FLLC
Sbjct: 346 GTAMRGKKNVKLWNHVLSKLSKSVPWIKSIEEKIFQP---LKLSYDFLE-DKAKFCFLLC 401
Query: 405 SVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVK 464
++FPED I V + R + G + E+ S E + NE L CLL D + VK
Sbjct: 402 ALFPEDYSIEVTEVVRYWMAEGFMEELGSQEDSMNEGITTVESLKDYCLLEDGDRRDTVK 461
Query: 465 MHDLVRNVAHWIAENE--------------IKCASEKDIMTLEHTSLRYLWCEKFPNSLD 510
MHD+VR+ A WI + +K +L SL E P+ ++
Sbjct: 462 MHDVVRDFAIWIMSSSQDDSHSLVMSGTGLQDIRQDKLAPSLRRVSLMNNKLESLPDLVE 521
Query: 511 --CSNLDFLQIH---TYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCI 565
C L + +V + LR+L L G + + SL L +L +
Sbjct: 522 EFCVKTSVLLLQGNFLLKEVPIGFLQAFPTLRILNL--SGTRIKSFPSCSLLRLFSLHSL 579
Query: 566 -LFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSE-CGMERNPFE 623
L + LV + + + KLE + LC +E P + +L R LDLS +E P
Sbjct: 580 FLRDCFKLVKLPSLETLAKLELLDLCGTHILEFPRGLEELKRFRHLDLSRTLHLESIPAR 639
Query: 624 VIARHTELEELFFADCRSKWEVE--------FLKEFSVPQVLQRYQIQLGS--------- 666
V++R + LE L +W V+ ++E Q LQ I+L S
Sbjct: 640 VVSRLSSLETLDMTSSHYRWSVQGETQKGQATVEEIGCLQRLQVLSIRLHSSPFLLNKRN 699
Query: 667 ----------MFSGFQDEFLNHH--RTLFLSYLDTSNAAIKDLAEKAEVLCI---AGIEG 711
+ G + H R L +S+L+ S +I L L + GIE
Sbjct: 700 TWIKRLKKFQLVVGSRYILRTRHDKRRLTISHLNVSQVSIGWLLAYTTSLALNHCQGIEA 759
Query: 712 GAKNIIPD--VFQSMNHL--KELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKH 767
K ++ D F+++ L + ++I + +E +V T + + L L H++
Sbjct: 760 MMKKLVSDNKGFKNLKSLTIENVIINTNSWVE-MVSTNTSKQSSDILDLLPNLEELHLRR 818
Query: 768 --LGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHI 825
L Q L E L+ + I+ C KL L + LE++++ C LQ++
Sbjct: 819 VDLETFSELQTHLGLKLETLKIIEITMCRKLRTLLDKRNFLTIPNLEEIEISYCDSLQNL 878
Score = 37.4 bits (85), Expect = 0.047
Identities = 26/116 (22%), Positives = 56/116 (47%), Gaps = 13/116 (11%)
Query: 949 MVSINTCMALHNNPRINEASHQTLQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCL 1007
++ I C L +++ + T+ N+ E+ ++ C+ L+ + + + P L
Sbjct: 840 IIEITMCRKLRT--LLDKRNFLTIPNLEEIEISYCDSLQNLHEALLY-----HQPFVPNL 892
Query: 1008 EMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLK 1063
+L L NLP L +C + +++ L+Q+E+ C +L C+ S G + ++K
Sbjct: 893 RVLKLRNLPNLVSIC-----NWGEVWECLEQVEVIHCNQLNCLPISSTCGRIKKIK 943
>At4g27220 putative protein
Length = 919
Score = 208 bits (529), Expect = 2e-53
Identities = 226/865 (26%), Positives = 396/865 (45%), Gaps = 96/865 (11%)
Query: 29 FNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQLLQM 88
F + + L + L ++ V + + R+ Q + + WL+ +NV L ++
Sbjct: 2 FRSNARALNRALERLKNVQTKVNEALKRSGIQEKSLERKLRIWLRKVE---ENVP-LGEL 57
Query: 89 AKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLSAGYFSAER 148
+++SC IW ++ +K + L+ E+G+ I+ S
Sbjct: 58 ILEKRSSCA------IWLSDKDVEILEKVKRLE---EQGQDLIKKISVNKSSREIVERVL 108
Query: 149 CWEFDSRKPAYE---ELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGK-----RCGNLFD 200
F +K A E +L LK +V IG++GMGG GKT L + F
Sbjct: 109 GPSFHPQKTALEMLDKLKDCLKKKNVQKIGVWGMGGVGKTTLVRTLNNDLLKYAATQQFA 168
Query: 201 QVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQM 260
V++V +S +++R+Q IA L F + +C RL L+ILDDVW
Sbjct: 169 LVIWVTVSKDFDLKRVQMDIAKRLGKRFTREQMNQLGLTICERLIDLKNFLLILDDVWHP 228
Query: 261 LDFDAIGIP-SIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALIS 319
+D D +GIP ++E K K+++TSR VC M + I+++ L E W+LF +
Sbjct: 229 IDLDQLGIPLALERSKDSKVVLTSRRLEVCQQMMTNENIKVACLQEKEAWELFCHN--VG 286
Query: 320 E-GTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVE-WKVALDRLRSSKP-VNIEK 376
E ++K +A+++S+EC GLP+A + + +L+GK +VE WK L+ L+ S P ++ E+
Sbjct: 287 EVANSDNVKPIAKDVSHECCGLPLAIITIGRTLRGKPQVEVWKHTLNLLKRSAPSIDTEE 346
Query: 377 GLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEG 436
+ + L+LSYD L + KS FL C++FPED I V L + G++ H YE
Sbjct: 347 KI---FGTLKLSYDFLQ-DNMKSCFLFCALFPEDYSIKVSELIMYWVAEGLLDGQHHYED 402
Query: 437 ARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENE--------------IK 482
NE +L SCLL D + VKMHD+VR+ A W ++ I+
Sbjct: 403 MMNEGVTLVERLKDSCLLEDGDSCDTVKMHDVVRDFAIWFMSSQGEGFHSLVMAGRGLIE 462
Query: 483 CASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD----FLQIHTYT-QVSDEIFKGMRMLR 537
+K + +++ SL E+ PN++ ++ LQ +++ +V + + LR
Sbjct: 463 FPQDKFVSSVQRVSLMANKLERLPNNV-IEGVETLVLLLQGNSHVKEVPNGFLQAFPNLR 521
Query: 538 VLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFVEL 597
+L L P ++L SL +L +L + L ++ + + KL+ + L + + EL
Sbjct: 522 ILDLSGVRIRTLPDSFSNLHSLRSL--VLRNCKKLRNLPSLESLVKLQFLDLHESAIREL 579
Query: 598 PDVVTQLTNLRLLDLSEC-GMERNPFEVIARHTELEELFFADCRSKWEVE--------FL 648
P + L++LR + +S ++ P I + + LE L A W ++ L
Sbjct: 580 PRGLEALSSLRYICVSNTYQLQSIPAGTILQLSSLEVLDMAGSAYSWGIKGEEREGQATL 639
Query: 649 KEFSVPQVLQRYQIQLGSMFS-GFQDEFLNHHRTLF---------------------LSY 686
E + LQ I+L + S ++ + L T F +S
Sbjct: 640 DEVTCLPHLQFLAIKLLDVLSFSYEFDSLTKRLTKFQFLFSPIRSVSPPGTGEGCLAISD 699
Query: 687 LDTSNAAIKDLAEKAEVLCI---AGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVD 743
++ SNA+I L + L + G+ G +N++ S +K L I + L
Sbjct: 700 VNVSNASIGWLLQHVTSLDLNYCEGLNGMFENLVTKSKSSFVAMKALSIHYFPSLS-LAS 758
Query: 744 TCLIEVGTLFFCKLHWLRIE--HMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFT 801
C ++ F L L ++ +++ +G L NG + + + L+ L +S C +L RLF+
Sbjct: 759 GCESQLD--LFPNLEELSLDNVNLESIGEL-NGFLGM--RLQKLKLLQVSGCRQLKRLFS 813
Query: 802 -LAVAQNLAQLEKLQVLSCPELQHI 825
+A L L++++V+SC L+ +
Sbjct: 814 DQILAGTLPNLQEIKVVSCLRLEEL 838
Score = 47.0 bits (110), Expect = 6e-05
Identities = 40/147 (27%), Positives = 72/147 (48%), Gaps = 27/147 (18%)
Query: 955 CMALHNNPRINEASHQTLQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCLEMLYLE 1013
C+A+ + N + LQ++T + +N CE L G+F+ + +T L + Y
Sbjct: 694 CLAISDVNVSNASIGWLLQHVTSLDLNYCEGLNGMFENL-VTKSKSSFVAMKALSIHYFP 752
Query: 1014 NLPQLRYLCKS-----------SVESTNL------------LFQNLQQMEISGCRRLKCI 1050
+L L C+S S+++ NL Q L+ +++SGCR+LK +
Sbjct: 753 SL-SLASGCESQLDLFPNLEELSLDNVNLESIGELNGFLGMRLQKLKLLQVSGCRQLKRL 811
Query: 1051 FS-SCMAGGLPQLKALKIEKCNQLDQI 1076
FS +AG LP L+ +K+ C +L+++
Sbjct: 812 FSDQILAGTLPNLQEIKVVSCLRLEEL 838
Score = 42.7 bits (99), Expect = 0.001
Identities = 26/79 (32%), Positives = 42/79 (52%), Gaps = 6/79 (7%)
Query: 971 TLQNITEVRVNNC-ELEGIFQLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVEST 1029
TL N+ E++V +C LE +F + D + L L ++ L+ LPQLR LC V
Sbjct: 820 TLPNLQEIKVVSCLRLEELFNFSSVPVDFCAESLLPKLTVIKLKYLPQLRSLCNDRV--- 876
Query: 1030 NLLFQNLQQMEISGCRRLK 1048
+ ++L+ +E+ C LK
Sbjct: 877 --VLESLEHLEVESCESLK 893
Score = 36.6 bits (83), Expect = 0.080
Identities = 34/136 (25%), Positives = 63/136 (46%), Gaps = 19/136 (13%)
Query: 956 MALHNNPRINEASHQTLQ-----NITEVRVNNCELEGIFQLVGLTNDGEKDPLTSCLEML 1010
+++H P ++ AS Q N+ E+ ++N LE I +L G + L++L
Sbjct: 746 LSIHYFPSLSLASGCESQLDLFPNLEELSLDNVNLESIGELNGFLGMRLQK-----LKLL 800
Query: 1011 YLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFS-------SCMAGGLPQLK 1063
+ QL+ L + + L NLQ++++ C RL+ +F+ C LP+L
Sbjct: 801 QVSGCRQLKRLFSDQILAGTL--PNLQEIKVVSCLRLEELFNFSSVPVDFCAESLLPKLT 858
Query: 1064 ALKIEKCNQLDQIVED 1079
+K++ QL + D
Sbjct: 859 VIKLKYLPQLRSLCND 874
>At1g61190
Length = 967
Score = 206 bits (524), Expect = 6e-53
Identities = 236/892 (26%), Positives = 400/892 (44%), Gaps = 102/892 (11%)
Query: 36 LAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQLLQMAKSE--K 93
L +E +L A + VQ++V R + + ++ E V+ WL N LL ++ E K
Sbjct: 38 LQREMEDLRATQHEVQNKVAREESRHQQRLEAVQVWLDRVNSIDIECKDLLSVSPVELQK 97
Query: 94 NSCFGHCPNWIWR-YSVGRKLSKKKRNLKLYIEEG----------RQYIEIERPASLSAG 142
G C ++ Y G+++ + EG R +E ERP + G
Sbjct: 98 LCLCGLCSKYVCSSYKYGKRVFLLLEEVTKLKSEGNFDEVSQPPPRSEVE-ERPTQPTIG 156
Query: 143 YFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKR---CGNLF 199
+ + K A+ LM +D V ++GL+GMGG GKT L ++ + G F
Sbjct: 157 --------QEEMLKKAWNRLM----EDGVGIMGLHGMGGVGKTTLFKKIHNKFAETGGTF 204
Query: 200 DQVLFVPISSTVEVERIQEKIAGSLEF---EFQEKDEMDRSKRLCMRLTQEDRVLVILDD 256
D V+++ +S ++ ++QE IA L ++ K+E D++ + R+ + R +++LDD
Sbjct: 205 DIVIWIVVSQGAKLSKLQEDIAEKLHLCDDLWKNKNESDKATDI-HRVLKGKRFVLMLDD 263
Query: 257 VWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQA 316
+W+ +D +AIGIP CK+ T+R + VC M K +Q+ L ++ W+LF+ +
Sbjct: 264 IWEKVDLEAIGIPYPSEVNKCKVAFTTRDQKVCGQMGDHKPMQVKCLEPEDAWELFKNK- 322
Query: 317 LISEGTWIS---IKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSKPV 372
+ + T S I +ARE++ +C+GLP+A + ++ K V EW+ A+D L S
Sbjct: 323 -VGDNTLRSDPVIVGLAREVAQKCRGLPLALSCIGETMASKTMVQEWEHAIDVLTRSAAE 381
Query: 373 NIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEV 431
+ +QN L+ SYD+L+ E KS FL C++FPED +I + L I G +GE
Sbjct: 382 FSD--MQNKILPILKYSYDSLEDEHIKSCFLYCALFPEDDKIDTKTLINKWICEGFIGED 439
Query: 432 HSYEGARNEVTVAKNKLISSCLLLDVNEGKCVK----MHDLVRNVAHWIA-------ENE 480
+ ARN+ LI + LL N+ VK MHD+VR +A WIA EN
Sbjct: 440 QVIKRARNKGYEMLGTLIRANLL--TNDRGFVKWHVVMHDVVREMALWIASDFGKQKENY 497
Query: 481 I--------KCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHTYTQVSDEIF 530
+ + KD + SL E+ CS L FLQ + +S E
Sbjct: 498 VVRARVGLHEIPKVKDWGAVRRMSLMMNEIEEITCESKCSELTTLFLQSNQLKNLSGEFI 557
Query: 531 KGMRMLRVLFL-YNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVG--DMKKLESI 587
+ M+ L VL L +N P + L SL L W ++ VG ++KKL +
Sbjct: 558 RYMQKLVVLDLSHNPDFNELPEQISGLVSLQYLDL----SWTRIEQLPVGLKELKKLIFL 613
Query: 588 TLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRSKWEVEF 647
LC + +++L +LR L L E + + V+ +LE L D R E
Sbjct: 614 NLCFTERLCSISGISRLLSLRWLSLRESNVHGDA-SVLKELQQLENL--QDLRITESAEL 670
Query: 648 LK-EFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLF-----LSYLDTSNAAIKDLAEKA 701
+ + + +++ +I+ G + F FL L+ SY N ++ ++
Sbjct: 671 ISLDQRLAKLISVLRIE-GFLQKPFDLSFLASMENLYGLLVENSYFSEINIKCRESETES 729
Query: 702 EVLCIAGIEGGAKNIIPDVFQSMNHLKELL------------IRDSKGI-ECLVDTCLIE 748
L I N+ + + +K+L IRDS+ + E + I
Sbjct: 730 SYLHINPKIPCFTNLTGLIIMKCHSMKDLTWILFAPNLVNLDIRDSREVGEIINKEKAIN 789
Query: 749 VGTLF--FCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQ 806
+ ++ F KL L + + L ++Y +P F L ++ + +CPKL +L A +
Sbjct: 790 LTSIITPFQKLERLFLYGLPKLESIYWSPLP----FPLLSNIVVKYCPKLRKLPLNATSV 845
Query: 807 NLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVL 858
L + +++ + PE ++ L +D D + + + + K H G L
Sbjct: 846 PLVEEFEIR-MDPPEQENELEWEDEDTKNRFLPSIKPLVRRLKIHYSGMGFL 896
>At1g61180
Length = 889
Score = 204 bits (520), Expect = 2e-52
Identities = 237/903 (26%), Positives = 408/903 (44%), Gaps = 126/903 (13%)
Query: 24 SYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVD 83
SY ++ L +E +L AI+ VQ++V R + + ++ E V+ WL N
Sbjct: 25 SYIRTLEKNLRALQREMEDLRAIQHEVQNKVARDEARHQRRLEAVQVWLDRVNSVDIECK 84
Query: 84 QLLQMAKSE--KNSCFGHCPNWIWR-YSVGRKLSKKKRNLKLYIEEG----------RQY 130
LL + E K G C ++ Y G+K+ +K EG R
Sbjct: 85 DLLSVTPVELQKLCLCGLCSKYVCSSYKYGKKVFLLLEEVKKLNSEGNFDEVSQPPPRSE 144
Query: 131 IEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAME 190
+E ERP + G + D + A+ LM +D V ++GL+GMGG GKT L +
Sbjct: 145 VE-ERPTQPTIG--------QEDMLEKAWNRLM----EDGVGIMGLHGMGGVGKTTLFKK 191
Query: 191 VGKR---CGNLFDQVLFVPISSTVEVERIQEKIAGSLEF---EFQEKDEMDRSKRLCMRL 244
+ + G FD V+++ +S V + ++QE IA L ++ K+E D++ + R+
Sbjct: 192 IHNKFAEIGGTFDIVIWIVVSKGVMISKLQEDIAEKLHLCDDLWKNKNESDKATDI-HRV 250
Query: 245 TQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLT 304
+ R +++LDD+W+ +D +AIGIP CK+ T+RS VC M K +Q++ L
Sbjct: 251 LKGKRFVLMLDDIWEKVDLEAIGIPYPSEVNKCKVAFTTRSREVCGEMGDHKPMQVNCLE 310
Query: 305 NDETWDLFQKQALISEGTWIS---IKNMAREISNECKGLPVATVAVASSLKGKAEV-EWK 360
++ W+LF+ + + + T S I +ARE++ +C+GLP+A + ++ K V EW+
Sbjct: 311 PEDAWELFKNK--VGDNTLSSDPVIVELAREVAQKCRGLPLALNVIGETMSSKTMVQEWE 368
Query: 361 VALDRLRSSKPVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLT 419
A+ +S + +QN L+ SYD+L E KS FL C++FPED EI E L
Sbjct: 369 HAIHVFNTSAAEFSD--MQNKILPILKYSYDSLGDEHIKSCFLYCALFPEDGEIYNEKLI 426
Query: 420 RSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIA-- 477
I G +GE + ARN+ L + LL V CV MHD+VR +A WIA
Sbjct: 427 DYWICEGFIGEDQVIKRARNKGYAMLGTLTRANLLTKVGTYYCV-MHDVVREMALWIASD 485
Query: 478 -----ENEI--------KCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHTY 522
EN + + KD + SL E+ CS L FLQ +
Sbjct: 486 FGKQKENFVVQAGVGLHEIPKVKDWGAVRKMSLMDNDIEEITCESKCSELTTLFLQSNKL 545
Query: 523 TQVSDEIFKGMRMLRVLFL-YNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDM 581
+ + M+ L VL L YN+ + P + L +L+ + S + + +
Sbjct: 546 KNLPGAFIRYMQKLVVLDLSYNRDFNKLP---EQISGLVSLQFLDLSNTSIEHMPI--GL 600
Query: 582 KKLESITLCDCSFVE-LPDV--VTQLTNLRLLDL--SECGMERNPFEVIARHTELEELFF 636
K+L+ +T D ++ + L + +++L +LRLL L S+ + + + + + L+EL
Sbjct: 601 KELKKLTFLDLTYTDRLCSISGISRLLSLRLLRLLGSKVHGDASVLKELQQLQNLQEL-- 658
Query: 637 ADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDE-FLNHHRTL-FLSYLDTSNAAI 694
+V L +L + S E FL L FL+ ++ ++
Sbjct: 659 -------------AITVSAELISLDQRLAKLISNLCIEGFLQKPFDLSFLASMENLSSLR 705
Query: 695 KDLAEKAEVLC-IAGIEGGAKNIIPDV--FQSMNHLK--------------------ELL 731
+ + +E+ C + E I P + F +++ L+ LL
Sbjct: 706 VENSYFSEIKCRESETESSYLRINPKIPCFTNLSRLEIMKCHSMKDLTWILFAPNLVVLL 765
Query: 732 IRDSKGIECLVD----TCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLED 787
I DS+ + +++ T L + F KL WL + ++ L ++Y +P F L
Sbjct: 766 IEDSREVGEIINKEKATNLTSITP--FLKLEWLILYNLPKLESIYWSPLP----FPVLLT 819
Query: 788 LYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAY-----DYRLL 842
+ +S+CPKL +L A + + + ++ + PE ++ L +D D + + Y+
Sbjct: 820 MDVSNCPKLRKLPLNATSVSKVEEFEIHMYPPPEQENELEWEDDDTKNRFLPSIKPYKYF 879
Query: 843 LFP 845
++P
Sbjct: 880 VYP 882
>At1g61310 similar to disease resistance protein
Length = 925
Score = 202 bits (515), Expect = 6e-52
Identities = 238/902 (26%), Positives = 391/902 (42%), Gaps = 145/902 (16%)
Query: 24 SYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVD 83
SY ++ L +E +L A + VQ++V R + + ++ E V+ WL N
Sbjct: 27 SYIRTLEKNLRALQREMEDLRATQHEVQNKVAREESRHQQRLEAVQVWLDRVNSIDIECK 86
Query: 84 QLLQMAKSE--KNSCFGHCPNWIWR-YSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLS 140
LL ++ E K G C ++ Y G+K+ +K+ EG + E+ +P S
Sbjct: 87 DLLSVSPVELQKLCLCGLCTKYVCSSYKYGKKVFLLLEEVKILKSEGN-FDEVSQPPPRS 145
Query: 141 AGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKR---CGN 197
++ E+ L +D V ++GL+GMGG GKT L ++ + G
Sbjct: 146 E--VEERPTQPTIGQEEMLEKAWNRLMEDGVGIMGLHGMGGVGKTTLFKKIHNKFAEIGG 203
Query: 198 LFDQVLFVPISSTVEVERIQEKIAGSLEF---EFQEKDEMDRSKRLCMRLTQEDRVLVIL 254
FD V+++ +S ++ ++QE IA L ++ K+E D++ + R+ + R +++L
Sbjct: 204 TFDIVIWIVVSQGAKLSKLQEDIAEKLHLCDDLWKNKNESDKATDI-HRVLKGKRFVLML 262
Query: 255 DDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQK 314
DD+W+ +D +AIGIP CK+ T+RS VC M K +Q++ L ++ W+LF+
Sbjct: 263 DDIWEKVDLEAIGIPYPSEVNKCKVAFTTRSREVCGEMGDHKPMQVNCLEPEDAWELFKN 322
Query: 315 QALISEGTWIS---IKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSK 370
+ + + T S I +ARE++ +C+GLP+A + ++ K V EW+ A+D L S
Sbjct: 323 K--VGDNTLSSDPVIVGLAREVAQKCRGLPLALNVIGETMASKTMVQEWEYAIDVLTRSA 380
Query: 371 PVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVG 429
G++N L+ SYD+L E KS FL C++FPED +I E L I G +G
Sbjct: 381 AE--FSGMENKILPILKYSYDSLGDEHIKSCFLYCALFPEDGQIYTETLIDKLICEGFIG 438
Query: 430 EVHSYEGARNE-----VTVAKNKLISSC------LLLDVNEGKCVKMHDLVRNVAHWIA- 477
E + ARN+ T+ + L++ LL V+ CV MHD+VR +A WIA
Sbjct: 439 EDQVIKRARNKGYAMLGTLTRANLLTKVGTELANLLTKVSIYHCV-MHDVVREMALWIAS 497
Query: 478 ------ENEIKCASE--------KDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHT 521
EN + AS KD + SL E+ CS L FLQ +
Sbjct: 498 DFGKQKENFVVQASAGLHEIPEVKDWGAVRRMSLMRNEIEEITCESKCSELTTLFLQSNQ 557
Query: 522 YTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDM 581
+S E + M+ L VL DL D
Sbjct: 558 LKNLSGEFIRYMQKLVVL-------------------------------DLSD------- 579
Query: 582 KKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRS 641
+ F ELP+ ++ L +L+ LDLS +E+ P + EL++L F D
Sbjct: 580 ---------NRDFNELPEQISGLVSLQYLDLSFTRIEQLPVGL----KELKKLTFLDL-- 624
Query: 642 KWEVEFLKEFSVPQVLQ-RYQIQLGSMFSGFQDEFLNHHRTLFLSYLD-TSNAAIKDLAE 699
+ + ++L R LGS G + L L T +A + L +
Sbjct: 625 AYTARLCSISGISRLLSLRVLSLLGSKVHGDASVLKELQQLENLQDLAITLSAELISLDQ 684
Query: 700 K-AEVLCIAGIEGGAKNIIPDVF-QSMNHLKELLIRDS--KGIECLVD------------ 743
+ A+V+ I GIEG + F SM +L L +++S I+C
Sbjct: 685 RLAKVISILGIEGFLQKPFDLSFLASMENLSSLWVKNSYFSEIKCRESETDSSYLHINPK 744
Query: 744 -TCLIEVGTLFFCKLH------W---------LRIEHMKHLGALYNGQMPLS----GHFE 783
C + L K H W L IE + +G + N + + F
Sbjct: 745 IPCFTNLSRLDIVKCHSMKDLTWILFAPNLVVLFIEDSREVGEIINKEKATNLTSITPFL 804
Query: 784 NLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLL 843
LE L + + PKL ++ + L L + V CP+L+ + + + ++R+L+
Sbjct: 805 KLERLILCYLPKLESIYWSPLPFPL--LLNIDVEECPKLRKLPL-NATSAPKVEEFRILM 861
Query: 844 FP 845
+P
Sbjct: 862 YP 863
>At1g12280 hypothetical protein
Length = 894
Score = 202 bits (515), Expect = 6e-52
Identities = 219/842 (26%), Positives = 384/842 (45%), Gaps = 82/842 (9%)
Query: 18 GVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAK-KQTRKTAEVVEKWLKDAN 76
G+ + Y C + V + K+ L RD V+ RV + + R+ V+ WL + +
Sbjct: 21 GLCINVGYICELSKNVVAMKKDMEVLKKKRDDVKRRVDIEEFTRRRERLSQVQGWLTNVS 80
Query: 77 IAMDNVDQLLQM--AKSEKNSCFGHCP-NWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEI 133
+ ++LL A+ ++ FG C N Y G+++ + ++ +G +
Sbjct: 81 TVENKFNELLTTNDAELQRLCLFGFCSKNVKMSYLYGKRVVLMLKEIESLSSQGD--FDT 138
Query: 134 ERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVG- 192
A+ A ++ E + L +D ++GLYGMGG GKT L +
Sbjct: 139 VTLATPIARIEEMPIQPTIVGQETMLERVWTRLTEDGDEIVGLYGMGGVGKTTLLTRINN 198
Query: 193 ---KRCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLC--MRLTQE 247
++C F V++V +S + ++ RIQ I L+ +E D ++ ++R + +
Sbjct: 199 KFSEKCSG-FGVVIWVVVSKSPDIHRIQGDIGKRLDLGGEEWDNVNENQRALDIYNVLGK 257
Query: 248 DRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDE 307
+ +++LDD+W+ ++ + +G+P GCK++ T+RS VC M +++S L +E
Sbjct: 258 QKFVLLLDDIWEKVNLEVLGVPYPSRQNGCKVVFTTRSRDVCGRMRVDDPMEVSCLEPNE 317
Query: 308 TWDLFQKQALISEGT---WISIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVAL 363
W+LFQ + + E T I +AR+++ +C GLP+A + ++ K V EW+ A+
Sbjct: 318 AWELFQMK--VGENTLKGHPDIPELARKVAGKCCGLPLALNVIGETMACKRMVQEWRNAI 375
Query: 364 DRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAI 423
D L S G++ L+ SYDNL+ E+ K FL CS+FPED + E L I
Sbjct: 376 DVLSSYAAE--FPGMEQILPILKYSYDNLNKEQVKPCFLYCSLFPEDYRMEKERLIDYWI 433
Query: 424 GLGIVGEVHSYEGARNEVTVAKNKLISSCLLLD--VNEGKCVKMHDLVRNVAHWIA---- 477
G + E S E A ++ L+ +CLLL+ +N+ + VKMHD+VR +A WIA
Sbjct: 434 CEGFIDENESRERALSQGYEIIGILVRACLLLEEAINKEQ-VKMHDVVREMALWIASDLG 492
Query: 478 ENEIKCASE-----------KDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIH-TYT 523
E++ +C + K+ ++ SL E S +C L FLQ + +
Sbjct: 493 EHKERCIVQVGVGLREVPKVKNWSSVRRMSLMENEIEILSGSPECLELTTLFLQKNDSLL 552
Query: 524 QVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVG--DM 581
+SDE F+ + ML VL L R+ L + L +LR + S W + VG ++
Sbjct: 553 HISDEFFRCIPMLVVLDLSGNSSLRK--LPNQISKLVSLRYLDLS-WTYIKRLPVGLQEL 609
Query: 582 KKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRS 641
KKL + L ++ ++ +++LR L L + M + ++ LE L +
Sbjct: 610 KKLRYLRLDYMKRLKSISGISNISSLRKLQLLQSKMSLD-MSLVEELQLLEHLEVLNISI 668
Query: 642 KWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKDLAEKA 701
K + K + P++++ QI + G Q+E L L +D N I
Sbjct: 669 KSSLVVEKLLNAPRLVKCLQI---LVLRGVQEE---SSGVLTLPDMDNLNKVIIRKCGMC 722
Query: 702 EVLCIAGIEGGAKNIIPDV----------FQSMNHLKELL------------IRDSKGIE 739
E+ + N P S + LK+L + DS+ +E
Sbjct: 723 EIKIERKTLSLSSNRSPKTQFLHNLSTVHISSCDGLKDLTWLLFAPNLTSLEVLDSELVE 782
Query: 740 CLV--DTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLT 797
++ + + G + F KL LR+ ++ L ++Y PLS F L+ ++I+ CP+L
Sbjct: 783 GIINQEKAMTMSGIIPFQKLESLRLHNLAMLRSIY--WQPLS--FPCLKTIHITKCPELR 838
Query: 798 RL 799
+L
Sbjct: 839 KL 840
>At1g12210 NBS/LRR disease resistance protein
Length = 885
Score = 192 bits (489), Expect = 7e-49
Identities = 230/897 (25%), Positives = 388/897 (42%), Gaps = 127/897 (14%)
Query: 27 CCFNNFVQDLAKEESNLA-------AIRDSVQDRVTRAK-KQTRKTAEVVEKWLKDANIA 78
C +++Q+L++ ++L A RD VQ R+ R + R+ V+ WL
Sbjct: 23 CVSGSYIQNLSENLASLQKAMGVLNAKRDDVQGRINREEFTGHRRRLAQVQVWLTRIQTI 82
Query: 79 MDNVDQLLQMAKSE-KNSCF-GHCP-NWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIER 135
+ + LL +E + C G C N Y G+++ R ++ +G +I
Sbjct: 83 ENQFNDLLSTCNAEIQRLCLCGFCSKNVKMSYLYGKRVIVLLREVEGLSSQG--VFDIVT 140
Query: 136 PASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRC 195
A+ A + +++ L +D V ++GLYGMGG GKT L ++ +
Sbjct: 141 EAAPIAEVEELPIQSTIVGQDSMLDKVWNCLMEDKVWIVGLYGMGGVGKTTLLTQINNKF 200
Query: 196 GNL---FDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLC--MRLTQEDRV 250
L FD V++V +S V +IQ+ I L + DE ++++R + + +
Sbjct: 201 SKLGGGFDVVIWVVVSKNATVHKIQKSIGEKLGLVGKNWDEKNKNQRALDIHNVLRRKKF 260
Query: 251 LVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWD 310
+++LDD+W+ ++ IG+P GCK+ T+ S+ VC M +++S L WD
Sbjct: 261 VLLLDDIWEKVELKVIGVPYPSGENGCKVAFTTHSKEVCGRMGVDNPMEISCLDTGNAWD 320
Query: 311 LFQKQALISEGTWIS---IKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRL 366
L +K+ + E T S I +AR++S +C GLP+A + ++ K + EW+ A + L
Sbjct: 321 LLKKK--VGENTLGSHPDIPQLARKVSEKCCGLPLALNVIGETMSFKRTIQEWRHATEVL 378
Query: 367 RSSKPVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGL 425
S+ + G+++ L+ SYD+L+ E+AKS FL CS+FPED EI E L I
Sbjct: 379 TSATDFS---GMEDEILPILKYSYDSLNGEDAKSCFLYCSLFPEDFEIRKEMLIEYWICE 435
Query: 426 GIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGK-CVKMHDLVRNVAHWI----AENE 480
G + E E A N+ L+ S LLL+ + K V MHD+VR +A WI +++
Sbjct: 436 GFIKEKQGREKAFNQGYDILGTLVRSSLLLEGAKDKDVVSMHDMVREMALWIFSDLGKHK 495
Query: 481 IKCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQIHTYTQVSDEIFKGMRMLRVLF 540
+C + I LD L E+ + R ++ +
Sbjct: 496 ERCIVQAGI-----------------------GLDEL---------PEV-ENWRAVKRMS 522
Query: 541 LYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDIS--FVGDMKKLESITLCD-CSFVEL 597
L N E+ + S + + + L + + LVDIS F M L + L + S EL
Sbjct: 523 LMNNNFEK---ILGSPECVELITLFLQNNYKLVDISMEFFRCMPSLAVLDLSENHSLSEL 579
Query: 598 PDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRSKWEVEFLKEFSVPQVL 657
P+ +++L +L+ LDLS +ER P + EL +L +E + S
Sbjct: 580 PEEISELVSLQYLDLSGTYIERLPHGL----HELRKLVHLKLERTRRLESISGIS----- 630
Query: 658 QRYQIQLGSMFSGFQDEFLNHHRTLFL----SYLDTSNAAIKDLAEKAEVLCIAGIEG-- 711
+L+ RTL L + LDT L E E++ G
Sbjct: 631 -----------------YLSSLRTLRLRDSKTTLDTGLMKELQLLEHLELITTDISSGLV 673
Query: 712 GAKNIIPDVFQSMNH--LKELLIRDSKGIECLV-----DTCLIEVGTLFFCKLHWLRIEH 764
G P V + + H +++ R + + LV + C I + + ++ +
Sbjct: 674 GELFCYPRVGRCIQHIYIRDHWERPEESVGVLVLPAIHNLCYISIWNCWMWEIMIEKTPW 733
Query: 765 MKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQH 824
K+L + +F NL ++ I C L L L A NL L+V C L+
Sbjct: 734 KKNL---------TNPNFSNLSNVRIEGCDGLKDLTWLLFAPNLI---NLRVWGCKHLED 781
Query: 825 ILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCN 881
I+ S + +L F KL+ ++ + L+ I L +L CL+I+ N
Sbjct: 782 II--SKEKAASVLEKEILPFQKLECLNLYQLSELKSIYWNALP--FQRLRCLDILNN 834
>At4g10780 putative disease resistance protein
Length = 892
Score = 192 bits (488), Expect = 9e-49
Identities = 239/919 (26%), Positives = 388/919 (42%), Gaps = 155/919 (16%)
Query: 9 SAISRDLVCGVIGQLSYPCCFN--NFVQDLA-------KEESNLAAIRDSVQDRVTRAKK 59
S IS + C + +Y C F+ N++ L K +L A RD V RV +
Sbjct: 3 SCISLQISCDQVLTRAYSCFFSLGNYIHKLKDNIVALEKAIEDLTATRDDVLRRVQMEEG 62
Query: 60 QTRKTAEVVEKWLKDANIAMDNVDQLLQMAKSE-KNSCF-GHC-PNWIWRYSVGRKLSKK 116
+ + + V+ WLK I + LL E + CF +C N Y+ G+++
Sbjct: 63 KGLERLQQVQVWLKRVEIIRNQFYDLLSARNIEIQRLCFYSNCSTNLSSSYTYGQRVFLM 122
Query: 117 KRNLKLYIEEGRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGL 176
+ ++ G I L R+ ++ L DD V +GL
Sbjct: 123 IKEVENLNSNGFFEIVAAPAPKLEMRPIQP----TIMGRETIFQRAWNRLMDDGVGTMGL 178
Query: 177 YGMGGCGKTMLAMEVGKR---CGNLFDQVLFVPISSTVEVERIQEKIAGSLEF---EFQE 230
YGMGG GKT L ++ N D V++V +SS +++ +IQE I L F E+ +
Sbjct: 179 YGMGGVGKTTLLTQIHNTLHDTKNGVDIVIWVVVSSDLQIHKIQEDIGEKLGFIGKEWNK 238
Query: 231 KDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCT 290
K E ++ + L+++ R +++LDD+W+ +D IGIPS CK++ T+RS VC
Sbjct: 239 KQESQKAVDILNCLSKK-RFVLLLDDIWKKVDLTKIGIPSQTRENKCKVVFTTRSLDVCA 297
Query: 291 LMDCQKKIQLSTLTNDETWDLFQ-KQALISEGTWISIKNMAREISNECKGLPVATVAVAS 349
M +++ L+ ++ W+LFQ K IS G+ I +A++++ +C+GLP+A +
Sbjct: 298 RMGVHDPMEVQCLSTNDAWELFQEKVGQISLGSHPDILELAKKVAGKCRGLPLALNVIGE 357
Query: 350 SLKGKAEV-EWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFP 408
++ GK V EW A+D L +S + L+ SYDNL+ + +S F C+++P
Sbjct: 358 TMAGKRAVQEWHHAVDVL-TSYAAEFSGMDDHILLILKYSYDNLNDKHVRSCFQYCALYP 416
Query: 409 EDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKC-VKMHD 467
ED I L I G + E A N+ L+ +CLL + + K VKMHD
Sbjct: 417 EDYSIKKYRLIDYWICEGFIDGNIGKERAVNQGYEILGTLVRACLLSEEGKNKLEVKMHD 476
Query: 468 LVRNVAHW----IAENEIKCASE-----------KDIMTLEHTSLRYLWCEKFPNSLDCS 512
+VR +A W + +N+ +C + +D + SL E+ S +C
Sbjct: 477 VVREMALWTLSDLGKNKERCIVQAGSGLRKVPKVEDWGAVRRLSLMNNGIEEISGSPECP 536
Query: 513 NLD--FLQIH-TYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSK 569
L FLQ + + +S E F+ MR L VL L
Sbjct: 537 ELTTLFLQENKSLVHISGEFFRHMRKLVVLDL---------------------------- 568
Query: 570 WDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHT 629
+ +L+ LP+ +++L LR LDLS +E P A
Sbjct: 569 ---------SENHQLDG----------LPEQISELVALRYLDLSHTNIEGLP----ACLQ 605
Query: 630 ELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLS---- 685
+L+ L + +E ++ +LGS+ +G L+ RTL L
Sbjct: 606 DLKTLIHLN------LECMR-------------RLGSI-AGISK--LSSLRTLGLRNSNI 643
Query: 686 YLDTSNAAIKDLAEKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLV--- 742
LD + L E E+L I + + D MN ++E+ IR CL+
Sbjct: 644 MLDVMSVKELHLLEHLEILTIDIVSTMVLEQMIDAGTLMNCMQEVSIR------CLIYDQ 697
Query: 743 --DTCL-------IEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHC 793
DT L + T++ C++ + IE + P S F NL + I C
Sbjct: 698 EQDTKLRLPTMDSLRSLTMWNCEISEIEIERLTW------NTNPTSPCFFNLSQVIIHVC 751
Query: 794 PKLTRLFTLAVAQNLA-----QLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLK 848
L L L A N+ QLE+LQ L + H +E ++++ F KL+
Sbjct: 752 SSLKDLTWLLFAPNITYLMIEQLEQLQEL----ISHAKATGVTEEEQQQLHKIIPFQKLQ 807
Query: 849 KFHVRECGVLEYIIPITLA 867
H+ L+ I I+L+
Sbjct: 808 ILHLSSLPELKSIYWISLS 826
>At5g63020 NBS/LRR disease resistance like protein
Length = 888
Score = 191 bits (484), Expect = 3e-48
Identities = 218/896 (24%), Positives = 396/896 (43%), Gaps = 131/896 (14%)
Query: 28 CFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAE----------VVEKWLKDANI 77
C N + E NL A++ +++ R + RK VV+ W+
Sbjct: 21 CLNRNGDYIHGLEENLTALQRALEQIEQRREDLLRKILSEERRGLQRLSVVQGWVSKVEA 80
Query: 78 AMDNVDQLLQMAKSE-KNSCF-GHCP-NWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIE 134
+ V++L++M + + C G C N + Y G+++ K +++ +G + E
Sbjct: 81 IVPRVNELVRMRSVQVQRLCLCGFCSKNLVSSYRYGKRVMKMIEEVEVLRYQGDFAVVAE 140
Query: 135 RPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKR 194
R + A + P E L +D++ ++GL+GMGG GKT L + R
Sbjct: 141 R---VDAARVEERPTRPMVAMDPMLESAWNRLMEDEIGILGLHGMGGVGKTTLLSHINNR 197
Query: 195 ---CGNLFDQVLFVPISSTVEVERIQEKIAGSLEFE---FQEKDEMDRSKRLCMRLTQED 248
G FD V+++ +S ++++RIQ++I L + +++K E ++ + + +
Sbjct: 198 FSRVGGEFDIVIWIVVSKELQIQRIQDEIWEKLRSDNEKWKQKTEDIKASNI-YNVLKHK 256
Query: 249 RVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDET 308
R +++LDD+W +D +G+P GCKI+ T+R + +C M +++ L D+
Sbjct: 257 RFVLLLDDIWSKVDLTEVGVPFPSRENGCKIVFTTRLKEICGRMGVDSDMEVRCLAPDDA 316
Query: 309 WDLFQKQ-ALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRL 366
WDLF K+ I+ G+ I +AR ++ +C+GLP+A + ++ K V EW+ A+D L
Sbjct: 317 WDLFTKKVGEITLGSHPEIPTVARTVAKKCRGLPLALNVIGETMAYKRTVQEWRSAIDVL 376
Query: 367 RSSKPVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGL 425
SS G+++ L+ SYDNL +E+ K F C++FPED I L IG
Sbjct: 377 TSSAAE--FSGMEDEILPILKYSYDNLKSEQLKLCFQYCALFPEDHNIEKNDLVDYWIGE 434
Query: 426 GIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIA-------E 478
G + + A N+ L+ SCLL++ N+ + VKMHD+VR +A WIA E
Sbjct: 435 GFID--RNKGKAENQGYEIIGILVRSCLLMEENQ-ETVKMHDVVREMALWIASDFGKQKE 491
Query: 479 NEIKCA--SEKDIMTLE------HTSLRYLWCEKFPNSLDCSNLD--FLQIHTYTQVSDE 528
N I A ++I +E SL + E ++ + L L+ + +S
Sbjct: 492 NFIVQAGLQSRNIPEIEKWKVARRVSLMFNNIESIRDAPESPQLITLLLRKNFLGHISSS 551
Query: 529 IFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDL-VDISFVGDMKKLESI 587
F+ M ML VL L + R+ R L + +L+ + S+ + + + + +++KL +
Sbjct: 552 FFRLMPMLVVLDL-SMNRDLRH-LPNEISECVSLQYLSLSRTRIRIWPAGLVELRKLLYL 609
Query: 588 TLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRSKWEVEF 647
L VE ++ LT+L++L L G +P
Sbjct: 610 NLEYTRMVESICGISGLTSLKVLRLFVSGFPEDPC------------------------V 645
Query: 648 LKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKDLAEKAEVLCIA 707
L E + + LQ I LG + ++FL++ R T I++L ++ V+
Sbjct: 646 LNELQLLENLQTLTITLG--LASILEQFLSNQRLASC----TRALRIENLNPQSSVISFV 699
Query: 708 GIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKH 767
+M+ L+EL DS E ++++ +
Sbjct: 700 A--------------TMDSLQELHFADSDIWE--------------------IKVKRNET 725
Query: 768 LGALYNGQMP-LSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHIL 826
+ L+ +P + F NL + + C +L L L A NL L+V+S +L+ ++
Sbjct: 726 VLPLH---IPTTTTFFPNLSQVSLEFCTRLRDLTWLIFAPNLT---VLRVISASDLKEVI 779
Query: 827 IDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYI----IPITLAQGLVQLECLEI 878
++ A L+ F +LK+ + +L++I +P Q ++ C E+
Sbjct: 780 -----NKEKAEQQNLIPFQELKELRLENVQMLKHIHRGPLPFPCLQKILVNGCSEL 830
>At4g26090 disease resistance protein RPS2
Length = 909
Score = 190 bits (482), Expect = 4e-48
Identities = 216/880 (24%), Positives = 378/880 (42%), Gaps = 118/880 (13%)
Query: 33 VQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQLLQMAKSE 92
+ DL +L AIRD + R+ + + R + +WL + LL +
Sbjct: 35 ITDLETAIGDLKAIRDDLTLRIQQDGLEGRSCSNRAREWLSAVQVTETKTALLLVRFRRR 94
Query: 93 KN---------SCFGHCPNWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLSAGY 143
+ SCFG C ++ KL KK + I E R+ E A + G
Sbjct: 95 EQRTRMRRRYLSCFG-CADY--------KLCKKVSAILKSIGELRERSE----AIKTDGG 141
Query: 144 FSAERCWEFDSRKPA-----YEELMCALKDDDVT-MIGLYGMGGCGKTMLAMEVGKRC-- 195
C E + E+++ L +++ +IG+YG GG GKT L +
Sbjct: 142 SIQVTCREIPIKSVVGNTTMMEQVLEFLSEEEERGIIGVYGPGGVGKTTLMQSINNELIT 201
Query: 196 -GNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDRVLVIL 254
G+ +D +++V +S IQ+ + L + EK+ + R ++ R L++L
Sbjct: 202 KGHQYDVLIWVQMSREFGECTIQQAVGARLGLSWDEKETGENRALKIYRALRQKRFLLLL 261
Query: 255 DDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLF-- 312
DDVW+ +D + G+P + CK++ T+RS A+C M + K+++ L W+LF
Sbjct: 262 DDVWEEIDLEKTGVPRPDRENKCKVMFTTRSIALCNNMGAEYKLRVEFLEKKHAWELFCS 321
Query: 313 ---QKQALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVALDRLRSS 369
+K L S SI+ +A I ++C GLP+A + + ++ + E + + +
Sbjct: 322 KVWRKDLLESS----SIRRLAEIIVSKCGGLPLALITLGGAMAHRETEEEWIHASEVLTR 377
Query: 370 KPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVG 429
P + KG+ + L+ SYDNL+++ +S FL C++FPE+ I +E L +G G +
Sbjct: 378 FPAEM-KGMNYVFALLKFSYDNLESDLLRSCFLYCALFPEEHSIEIEQLVEYWVGEGFLT 436
Query: 430 EVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENEIKCASEKDI 489
H + + L ++CLL +E VKMH++VR+ A W+A + + K++
Sbjct: 437 SSHGVNTIYKGYFLIGD-LKAACLLETGDEKTQVKMHNVVRSFALWMASEQ---GTYKEL 492
Query: 490 MTLE----HT--------------SLRYLWCEKFPNSLDCSNLDFLQIH---TYTQVSDE 528
+ +E HT SL + P L C L L + + ++
Sbjct: 493 ILVEPSMGHTEAPKAENWRQALVISLLDNRIQTLPEKLICPKLTTLMLQQNSSLKKIPTG 552
Query: 529 IFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDI-SFVGDMKKLESI 587
F M +LRVL L PL S+K L L + S + + +G+++KL+ +
Sbjct: 553 FFMHMPVLRVLDLSFTSITEIPL---SIKYLVELYHLSMSGTKISVLPQELGNLRKLKHL 609
Query: 588 TLCDCSFVE-LP-DVVTQLTNLRLLDL--SECGMERNPFEVIARHTELEELFFADCRSKW 643
L F++ +P D + L+ L +L+L S G E F E EEL FAD
Sbjct: 610 DLQRTQFLQTIPRDAICWLSKLEVLNLYYSYAGWELQSFG----EDEAEELGFAD----- 660
Query: 644 EVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKDLAEKAEV 703
+E+L+ + + L ++F EF H+ + +++ N E+
Sbjct: 661 -LEYLENLTTLGITVLSLETLKTLF-----EFGALHKHIQHLHVEECN----------EL 704
Query: 704 LCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIE 763
L +P + +L+ L I+ +E LV E L L L +
Sbjct: 705 LYFN---------LPSLTNHGRNLRRLSIKSCHDLEYLVTPADFENDWL--PSLEVLTLH 753
Query: 764 HMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQ 823
+ +L ++ G N+ + ISHC KL + + Q L +LE +++ C E++
Sbjct: 754 SLHNLTRVW-GNSVSQDCLRNIRCINISHCNKLKNV---SWVQKLPKLEVIELFDCREIE 809
Query: 824 HILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIP 863
++ + + + LFP LK R+ L I+P
Sbjct: 810 ELISEHESPSVEDPT----LFPSLKTLRTRDLPELNSILP 845
Score = 35.8 bits (81), Expect = 0.14
Identities = 59/264 (22%), Positives = 115/264 (43%), Gaps = 36/264 (13%)
Query: 821 ELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVC 880
EL H+ + + IS L KLK ++ L+ IP L +LE L
Sbjct: 582 ELYHLSMSGTK--ISVLPQELGNLRKLKHLDLQRTQFLQ-TIPRDAICWLSKLEVL---- 634
Query: 881 NENLKYVF-GQSTHNDGQNQ-NELKIIELSALEELTLVNLPNINSICPEDCY---LMWPS 935
NL Y + G + G+++ EL +L LE LT + + ++ + + +
Sbjct: 635 --NLYYSYAGWELQSFGEDEAEELGFADLEYLENLTTLGITVLSLETLKTLFEFGALHKH 692
Query: 936 LLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVRVNNCELEGIFQLVGLT 995
+ +++ C E ++ + L N+ R N+ + + +C + +
Sbjct: 693 IQHLHVEECNELLYFNLPS---LTNHGR----------NLRRLSIKSCH---DLEYLVTP 736
Query: 996 NDGEKDPLTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCM 1055
D E D L S LE+L L +L L + +SV L +N++ + IS C +LK +
Sbjct: 737 ADFENDWLPS-LEVLTLHSLHNLTRVWGNSVSQDCL--RNIRCINISHCNKLKNV---SW 790
Query: 1056 AGGLPQLKALKIEKCNQLDQIVED 1079
LP+L+ +++ C ++++++ +
Sbjct: 791 VQKLPKLEVIELFDCREIEELISE 814
>At1g15890 hypothetical protein
Length = 851
Score = 185 bits (469), Expect = 1e-46
Identities = 179/622 (28%), Positives = 289/622 (45%), Gaps = 58/622 (9%)
Query: 36 LAKEESNLAAIRDSVQDRVTRAKKQTRKTA----------EVVEKWLKDANIAMDNVDQL 85
+ K E+NL A+++++Q+ R R+ V+ WL V+ L
Sbjct: 29 ILKMEANLEALQNTMQELEERRDDLLRRVVIEEDKGLQRLAQVQGWLSRVKDVCSQVNDL 88
Query: 86 LQMA--KSEKNSCFGHCP-NWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLSAG 142
L+ ++E+ G+C N+I + G + KK ++++ + +G + E+ +
Sbjct: 89 LKAKSIQTERLCLCGYCSKNFISGRNYGINVLKKLKHVEGLLAKGVFEVVAEKIPAPKVE 148
Query: 143 YFSAERCWEFDSRKP-AYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRC---GNL 198
+ D+ A+ LM D+ +GLYGMGG GKT L + + N
Sbjct: 149 KKHIQTTVGLDAMVGRAWNSLM----KDERRTLGLYGMGGVGKTTLLASINNKFLEGMNG 204
Query: 199 FDQVLFVPISSTVEVERIQEKIAGSLEFE--FQEKDEMDRSKRLCMRLTQEDRVLVILDD 256
FD V++V +S ++ E IQE+I G L +++ E +++ +C L + VL +LDD
Sbjct: 205 FDLVIWVVVSKDLQNEGIQEQILGRLGLHRGWKQVTEKEKASYICNILNVKKFVL-LLDD 263
Query: 257 VWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQA 316
+W +D + IG+P + G KI+ T+RS+ VC M+ ++++ L DE W+LFQK+
Sbjct: 264 LWSEVDLEKIGVPPLTRENGSKIVFTTRSKDVCRDMEVDGEMKVDCLPPDEAWELFQKKV 323
Query: 317 -LISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSK---P 371
I + I +AR+++ +C GLP+A + ++ + V EW+ + L SS P
Sbjct: 324 GPIPLQSHEDIPTLARKVAEKCCGLPLALSVIGKAMASRETVQEWQHVIHVLNSSSHEFP 383
Query: 372 VNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEV 431
EK L L+ SYD+L E+ K FL CS+FPED E+ E L + G +
Sbjct: 384 SMEEKILP----VLKFSYDDLKDEKVKLCFLYCSLFPEDYEVRKEELIEYWMCEGFIDGN 439
Query: 432 HSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENEIK--------- 482
+GA N+ L+ + LL+D VKMHD++R +A WIA N K
Sbjct: 440 EDEDGANNKGHDIIGSLVRAHLLMDGELTTKVKMHDVIREMALWIASNFGKQKETLCVKP 499
Query: 483 ----CASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD-----FLQIHTYTQVSDEIFKGM 533
C KDI E L C + N SN LQ + +S + F+ M
Sbjct: 500 GVQLCHIPKDI-NWESLRRMSLMCNQIANISSSSNSPNLSTLLLQNNKLVHISCDFFRFM 558
Query: 534 RMLRVLFL-YNKGRERRPLLTTSLKSLTNLRCILFS-KWDLVDISFVGDMKKLESITLCD 591
L VL L N P + L SL + KW + +SF ++KKL + L
Sbjct: 559 PALVVLDLSRNSSLSSLPEAISKLGSLQYINLSTTGIKW--LPVSF-KELKKLIHLNLEF 615
Query: 592 CSFVE-LPDVVTQLTNLRLLDL 612
+E + + T L NL++L L
Sbjct: 616 TDELESIVGIATSLPNLQVLKL 637
>At1g12220 NBS/LRR disease resistance protein, putative
Length = 889
Score = 182 bits (462), Expect = 9e-46
Identities = 179/637 (28%), Positives = 294/637 (46%), Gaps = 49/637 (7%)
Query: 19 VIGQLSYPCC-----FNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQT----RKTAEVVE 69
V+ Q S C +N ++LA + + ++ D + R + + ++ V+
Sbjct: 14 VVSQFSQLLCVRGSYIHNLSKNLASLQKAMRMLKARQYDVIRRLETEEFTGRQQRLSQVQ 73
Query: 70 KWLKDANIAMDNVDQLLQMAKSE-KNSCF-GHCPNWI-WRYSVGRKLSKKKRNLKLYIEE 126
WL I + + LL+ + E + C G C + Y G+++ + ++ +
Sbjct: 74 VWLTSVLIIQNQFNDLLRSNEVELQRLCLCGFCSKDLKLSYRYGKRVIMMLKEVESLSSQ 133
Query: 127 GRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTM 186
G + ++ A+ A ++ E+ L +D ++GLYGMGG GKT
Sbjct: 134 G--FFDVVSEATPFADVDEIPFQPTIVGQEIMLEKAWNRLMEDGSGILGLYGMGGVGKTT 191
Query: 187 LAMEVGKRCGNL---FDQVLFVPISSTVEVERIQEKIA---GSLEFEFQEKDEMDRSKRL 240
L ++ + + FD V++V +S + V +IQ IA G E+ EK++ + +
Sbjct: 192 LLTKINNKFSKIDDRFDVVIWVVVSRSSTVRKIQRDIAEKVGLGGMEWSEKNDNQIAVDI 251
Query: 241 CMRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQL 300
L + VL +LDD+W+ ++ A+G+P GCK+ T+RS VC M +++
Sbjct: 252 HNVLRRRKFVL-LLDDIWEKVNLKAVGVPYPSKDNGCKVAFTTRSRDVCGRMGVDDPMEV 310
Query: 301 STLTNDETWDLFQ-KQALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEV-E 358
S L +E+WDLFQ K + G+ I +AR+++ +C+GLP+A + ++ K V E
Sbjct: 311 SCLQPEESWDLFQMKVGKNTLGSHPDIPGLARKVARKCRGLPLALNVIGEAMACKRTVHE 370
Query: 359 WKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFL 418
W A+D L SS ++ L+ SYDNL+ E KS FL CS+FPED I E L
Sbjct: 371 WCHAIDVLTSS-AIDFSGMEDEILHVLKYSYDNLNGELMKSCFLYCSLFPEDYLIDKEGL 429
Query: 419 TRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKC-VKMHDLVRNVAHWIA 477
I G + E E N+ L+ +CLLL+ K VKMHD+VR +A WI+
Sbjct: 430 VDYWISEGFINEKEGRERNINQGYEIIGTLVRACLLLEEERNKSNVKMHDVVREMALWIS 489
Query: 478 ----ENEIKC-----------ASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIH 520
+ + KC KD T+ SL E+ +S +C+ L FLQ +
Sbjct: 490 SDLGKQKEKCIVRAGVGLREVPKVKDWNTVRKISLMNNEIEEIFDSHECAALTTLFLQKN 549
Query: 521 TYTQVSDEIFKGMRMLRVLFLYNKGRERRPL--LTTSLKSLTNLRCILFSKWDLVDISF- 577
++S E F+ M L VL L E + L L + L +LR S + +
Sbjct: 550 DVVKISAEFFRCMPHLVVLDL----SENQSLNELPEEISELASLRYFNLSYTCIHQLPVG 605
Query: 578 VGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSE 614
+ +KKL + L S + ++ L NLR L L +
Sbjct: 606 LWTLKKLIHLNLEHMSSLGSILGISNLWNLRTLGLRD 642
>At1g61300 similar to disease resistance protein
Length = 763
Score = 180 bits (456), Expect = 4e-45
Identities = 191/697 (27%), Positives = 323/697 (45%), Gaps = 70/697 (10%)
Query: 166 LKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGNL---FDQVLFVPISSTVEVERIQEKIAG 222
L +D V ++GL+GMGG GKT L ++ + + FD V+++ +S ++ ++QE IA
Sbjct: 56 LMEDRVGIMGLHGMGGVGKTTLFKKIHNKFAKMSSRFDIVIWIVVSKGAKLSKLQEDIAE 115
Query: 223 SLEF---EFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKI 279
L ++ K+E D++ + R+ + R +++LDD+W+ +D +AIG+P CK+
Sbjct: 116 KLHLCDDLWKNKNESDKATDI-HRVLKGKRFVLMLDDIWEKVDLEAIGVPYPSEVNKCKV 174
Query: 280 LITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALISEGTWIS---IKNMAREISNE 336
T+R + VC M K +Q+ L ++ W+LF+ + + + T S I +ARE++ +
Sbjct: 175 AFTTRDQKVCGEMGDHKPMQVKCLEPEDAWELFKNK--VGDNTLRSDPVIVELAREVAQK 232
Query: 337 CKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTE 395
C+GLP+A + ++ K V EW+ A+D L S G L+ SYD+L E
Sbjct: 233 CRGLPLALSVIGETMASKTMVQEWEHAIDVLTRSAAEFSNMG-NKILPILKYSYDSLGDE 291
Query: 396 EAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLL 455
KS FL C++FPED EI E L I G +GE + ARN+ L + LL
Sbjct: 292 HIKSCFLYCALFPEDDEIYNEKLIDYWICEGFIGEDQVIKRARNKGYEMLGTLTLANLLT 351
Query: 456 DVNEGKCVKMHDLVRNVAHWIA-------ENEIKCA--------SEKDIMTLEHTSLRYL 500
V + V MHD+VR +A WIA EN + A KD + SL
Sbjct: 352 KVGT-EHVVMHDVVREMALWIASDFGKQKENFVVRARVGLHERPEAKDWGAVRRMSLMDN 410
Query: 501 WCEKFPNSLDCSNLD--FLQIHTYTQVSDEIFKGMRMLRVLFL-YNKGRERRPLLTTSLK 557
E+ CS L FLQ + +S E + M+ L VL L YN+ + P +
Sbjct: 411 HIEEITCESKCSELTTLFLQSNQLKNLSGEFIRYMQKLVVLDLSYNRDFNKLP---EQIS 467
Query: 558 SLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSF-VELPDV--VTQLTNLRLLDL-- 612
L +L+ + S + + VG +KKL+ +T + ++ V L + +++L +LRLL L
Sbjct: 468 GLVSLQFLDLSNTSIKQLP-VG-LKKLKKLTFLNLAYTVRLCSISGISRLLSLRLLRLLG 525
Query: 613 SECGMERNPFEVIARHTELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQ 672
S+ + + + + + L+ L + E S+ Q L LG GF
Sbjct: 526 SKVHGDASVLKELQKLQNLQHL---------AITLSAELSLNQRLANLISILG--IEGFL 574
Query: 673 DEFLNHHRTLFLSYLDTSNAAIKDLAEKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLI 732
+ + FL+ ++ ++ + +E+ C + I +L L +
Sbjct: 575 QKPFD---LSFLASMENLSSLWVKNSYFSEIKCRESETASSYLRINPKIPCFTNLSRLGL 631
Query: 733 RDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLS----GHFENLEDL 788
I+ L LF L +L IE + +G + N + + F LE L
Sbjct: 632 SKCHSIKDL-------TWILFAPNLVYLYIEDSREVGEIINKEKATNLTSITPFLKLERL 684
Query: 789 YISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHI 825
+ + PKL ++ + + +L + VL CP+L+ +
Sbjct: 685 ILYNLPKLESIYWSPL--HFPRLLIIHVLDCPKLRKL 719
>At1g12290 hypothetical protein
Length = 884
Score = 178 bits (452), Expect = 1e-44
Identities = 213/828 (25%), Positives = 371/828 (44%), Gaps = 95/828 (11%)
Query: 36 LAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQL--LQMAKSEK 93
L + +L A+RD + +V A++ + ++ WLK + L + + ++
Sbjct: 39 LEEAMEDLKALRDDLLRKVQTAEEGGLQRLHQIKVWLKRVKTIESQFNDLDSSRTVELQR 98
Query: 94 NSCFG-HCPNWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLSAGYFSAERCWE- 151
C G N Y GR++ N+ ++ + E+ PA+ + G ER +
Sbjct: 99 LCCCGVGSRNLRLSYDYGRRVFLML-NIVEDLKSKGIFEEVAHPATRAVG---EERPLQP 154
Query: 152 -FDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGNLFDQV---LFVPI 207
++ E+ L DD ++GLYGMGG GKT L ++ R + D V ++V +
Sbjct: 155 TIVGQETILEKAWDHLMDDGTKIMGLYGMGGVGKTTLLTQINNRFCDTDDGVEIVIWVVV 214
Query: 208 SSTVEVERIQEKIAGSLEF---EFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFD 264
S +++ +IQ++I + F E+ +K E ++ + L+++ R +++LDD+W+ ++
Sbjct: 215 SGDLQIHKIQKEIGEKIGFIGVEWNQKSENQKAVDILNFLSKK-RFVLLLDDIWKRVELT 273
Query: 265 AIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQ-ALISEGTW 323
IGIP+ GCKI T+R ++VC M +++ L D+ WDLF+K+ I+ +
Sbjct: 274 EIGIPNPTSENGCKIAFTTRCQSVCASMGVHDPMEVRCLGADDAWDLFKKKVGDITLSSH 333
Query: 324 ISIKNMAREISNECKGLPVATVAVASSLK-GKAEVEWKVALDRLRSSKPVNIEKGLQNPY 382
I +AR+++ C GLP+A + ++ K EW A+D + ++ N +
Sbjct: 334 PDIPEIARKVAQACCGLPLALNVIGETMACKKTTQEWDRAVD-VSTTYAANFGAVKERIL 392
Query: 383 KCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVT 442
L+ SYDNL++E K+ FL CS+FPED I E L I G + + +GA E
Sbjct: 393 PILKYSYDNLESESVKTCFLYCSLFPEDDLIEKERLIDYWICEGFIDGDENKKGAVGEGY 452
Query: 443 VAKNKLISSCLLLD---VNEGKCVKMHDLVRNVAHWIAE-----------------NEIK 482
L+ + LL++ N VKMHD+VR +A WIA NEI
Sbjct: 453 EILGTLVCASLLVEGGKFNNKSYVKMHDVVREMALWIASDLRKHKDNCIVRAGFRLNEI- 511
Query: 483 CASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHTY-TQVSDEIFKGMRMLRVL 539
KD + SL ++ S +C L FLQ + + +S E F+ M L VL
Sbjct: 512 -PKVKDWKVVSRMSLVNNRIKEIHGSPECPKLTTLFLQDNRHLVNISGEFFRSMPRLVVL 570
Query: 540 FL-YNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFVELP 598
L +N P + L SL L + +S + + + +KKL + L +E
Sbjct: 571 DLSWNVNLSGLPDQISELVSLRYLD-LSYSSIGRLPVGLL-KLKKLMHLNLESMLCLESV 628
Query: 599 DVVTQLTNL---RLLDLSEC-------------GMERNPFEVIARHTELEELF----FAD 638
+ L+NL RLL+L +E E+I+ + LE+L
Sbjct: 629 SGIDHLSNLKTVRLLNLRMWLTISLLEELERLENLEVLTIEIIS-SSALEQLLCSHRLVR 687
Query: 639 CRSKWEVEFLKEFSV-----PQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAA 693
C K V++L E SV P + ++ +G G +D + + +L ++
Sbjct: 688 CLQKVSVKYLDEESVRILTLPSIGDLREVFIGG--CGMRDIIIERNTSL-------TSPC 738
Query: 694 IKDLAEKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLF 753
+L++ C G K++ +F +L L + +S+ IE ++ +
Sbjct: 739 FPNLSKVLITGC-----NGLKDLTWLLFAP--NLTHLNVWNSRQIEEIISQEKASTADIV 791
Query: 754 -FCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISH-CPKLTRL 799
F KL +L + + L ++Y +P F L + + + C KLT+L
Sbjct: 792 PFRKLEYLHLWDLPELKSIYWNPLP----FPCLNQINVQNKCRKLTKL 835
>At5g43730 disease resistance protein
Length = 848
Score = 170 bits (431), Expect = 4e-42
Identities = 177/633 (27%), Positives = 282/633 (43%), Gaps = 65/633 (10%)
Query: 28 CFNNFVQDLAKEESNLAAI----------RDSVQDRVTRAKKQTRKTAEVVEKWLKDANI 77
CF + + ESNL A+ RD + RV+ + + + +V WL I
Sbjct: 20 CFLSDSNYIHLMESNLDALQKTMEELKNGRDDLLARVSIEEDKGLQRLALVNGWLSRVQI 79
Query: 78 AMDNVDQLLQMAKSEKNSC--FGHCP-NWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIE 134
LL+ E FG+C + I Y+ G K+ K +K + + + + +
Sbjct: 80 VESEFKDLLEAMSIETGRLCLFGYCSEDCISSYNYGGKVMKNLEEVKELLSK-KNFEVVA 138
Query: 135 RPASLSAGYFSAERCWEFDSRKP-AYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGK 193
+ A + D+ A+E L+ DD++ +GLYGMGG GKT L +
Sbjct: 139 QKIIPKAEKKHIQTTVGLDTMVGIAWESLI----DDEIRTLGLYGMGGIGKTTLLESLNN 194
Query: 194 RCGNL---FDQVLFVPISSTVEVERIQEKIAGSL--EFEFQEKDEMDRSKRLCMRLTQED 248
+ L FD V++V +S ++E IQ++I G L + E++ + E ++ + L ++
Sbjct: 195 KFVELESEFDVVIWVVVSKDFQLEGIQDQILGRLRPDKEWERETESKKASLINNNLKRKK 254
Query: 249 RVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDET 308
VL +LDD+W +D IG+P G KI+ T+RS+ VC M K+I++ L+ DE
Sbjct: 255 FVL-LLDDLWSEVDLIKIGVPPPSRENGSKIVFTTRSKEVCKHMKADKQIKVDCLSPDEA 313
Query: 309 WDLFQKQALISEGTWI-----SIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVA 362
W+LF+ ++ G I I +AR ++ +C GLP+A + ++ K V EW+ A
Sbjct: 314 WELFR----LTVGDIILRSHQDIPALARIVAAKCHGLPLALNVIGKAMVCKETVQEWRHA 369
Query: 363 LDRLRSSKPVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRS 421
++ L S P + G++ L+ SYD+L E K FL CS+FPED EI + L
Sbjct: 370 INVLNS--PGHKFPGMEERILPILKFSYDSLKNGEIKLCFLYCSLFPEDFEIEKDKLIEY 427
Query: 422 AIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWI----- 476
I G + +G N+ L+ + LL++ VKMHD++R +A WI
Sbjct: 428 WICEGYINPNRYEDGGTNQGYDIIGLLVRAHLLIECELTDKVKMHDVIREMALWINSDFG 487
Query: 477 AENEIKCASE--------KDIM--TLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHTYTQ 524
+ E C DI + SL EK S +C NL L +
Sbjct: 488 NQQETICVKSGAHVRLIPNDISWEIVRQMSLISTQVEKIACSPNCPNLSTLLLPYNKLVD 547
Query: 525 VSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDI-SFVGDMKK 583
+S F M L VL L L + ++NL + + L I S +KK
Sbjct: 548 ISVGFFLFMPKLVVLDLSTNWS-----LIELPEEISNLGSLQYLNLSLTGIKSLPVGLKK 602
Query: 584 LESITLCDCSFV----ELPDVVTQLTNLRLLDL 612
L + + F L + T L NL++L L
Sbjct: 603 LRKLIYLNLEFTNVLESLVGIATTLPNLQVLKL 635
>At5g47250 NBS/LRR disease resistance protein
Length = 843
Score = 170 bits (430), Expect = 5e-42
Identities = 214/880 (24%), Positives = 366/880 (41%), Gaps = 143/880 (16%)
Query: 7 FASAISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAE 66
+ SA+S +C +G + C + L L A ++ V +RV + + +
Sbjct: 12 YKSALS--YLCVKVGNI---CMLKENLVLLKSAFDELKAEKEDVVNRVNAGELKGGQRLA 66
Query: 67 VVEKWLKDANIAMDNVDQLLQMAKSEKNSC---------FGHCPNWIWRYSVGRKLSKKK 117
+V WL I +N QL+ +A + S W ++G K+ KK
Sbjct: 67 IVATWLSQVEIIEENTKQLMDVASARDASSQNASAVRRRLSTSGCWFSTCNLGEKVFKKL 126
Query: 118 RNLKLYIEEGRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLY 177
+K + Q + + P + C + E+ +L+ D+ M+G++
Sbjct: 127 TEVKSLSGKDFQEVTEQPPPPV----VEVRLCQQTVGLDTTLEKTWESLRKDENRMLGIF 182
Query: 178 GMGGCGKTMLAMEVGKRCGNL---FDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEM 234
GMGG GKT L + + + +D V++V S +V +IQ+ I L
Sbjct: 183 GMGGVGKTTLLTLINNKFVEVSDDYDVVIWVESSKDADVGKIQDAIGERLHICDNNWSTY 242
Query: 235 DRSKRLC----MRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCT 290
R K+ + + R +++LDD+W+ + AIGIP + K K++ T+RS+ VC+
Sbjct: 243 SRGKKASEISRVLRDMKPRFVLLLDDLWEDVSLTAIGIPVLG--KKYKVVFTTRSKDVCS 300
Query: 291 LMDCQKKIQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVATVAVASS 350
+M + I++ L+ ++ WDLF + IS ++A++I +C GLP+A + +
Sbjct: 301 VMRANEDIEVQCLSENDAWDLFDMKVHCDGLNEIS--DIAKKIVAKCCGLPLALEVIRKT 358
Query: 351 LKGKAEV-EWKVALDRLRS--SKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVF 407
+ K+ V +W+ ALD L S S+ EKG+ ++ L+LSYD L T+ AK FL C++F
Sbjct: 359 MASKSTVIQWRRALDTLESYRSEMKGTEKGI---FQVLKLSYDYLKTKNAK-CFLYCALF 414
Query: 408 PEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHD 467
P+ I + L IG G + E E A++ + L+ + LLL+ N K V MHD
Sbjct: 415 PKAYYIKQDELVEYWIGEGFIDEKDGRERAKDRGYEIIDNLVGAGLLLESN--KKVYMHD 472
Query: 468 LVRNVAHWIAENEIKCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQI---HTYTQ 524
++R++A WI +E + + T S + P+ D + + + +
Sbjct: 473 MIRDMALWIV-SEFRDGERYVVKTDAGLS-------QLPDVTDWTTVTKMSLFNNEIKNI 524
Query: 525 VSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKL 584
D F L LFL N L + ++ S ++D+S+
Sbjct: 525 PDDPEFPDQTNLVTLFLQN----------NRLVDIVGKFFLVMSTLVVLDLSW------- 567
Query: 585 ESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRSKWE 644
+ ELP ++ L +LRLL+LS ++ P E + ++L L +
Sbjct: 568 ------NFQITELPKGISALVSLRLLNLSGTSIKHLP-EGLGVLSKLIHLNLESTSNLRS 620
Query: 645 VEFLKEFSVPQVLQRY---------QIQLGSMFSGFQ------------DEFLNHHRT-- 681
V + E QVL+ Y +++ G Q +EFL R
Sbjct: 621 VGLISELQKLQVLRFYGSAAALDCCLLKILEQLKGLQLLTVTVNNDSVLEEFLGSTRLAG 680
Query: 682 ----LFLSYLDTSNAAIKDLA--EKAEVLCIAGIEGGA---------------------- 713
++L L S AAI L+ K E++ E G
Sbjct: 681 MTQGIYLEGLKVSFAAIGTLSSLHKLEMVNCDITESGTEWEGKRRDQYSPSTSSSEITPS 740
Query: 714 ----KNIIPDVFQSMNHLKELL------------IRDSKGIECLVDTCLIE-VGTLFFCK 756
K++ V S HLK+L + S + L++ + VG F +
Sbjct: 741 NPWFKDLSAVVINSCIHLKDLTWLMYAANLESLSVESSPKMTELINKEKAQGVGVDPFQE 800
Query: 757 LHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKL 796
L LR+ ++K LG++Y Q+ N D I +CP L
Sbjct: 801 LQVLRLHYLKELGSIYGSQVSFPKLKLNKVD--IENCPNL 838
>At1g63360 disease resistance protein, putative
Length = 884
Score = 168 bits (425), Expect = 2e-41
Identities = 220/861 (25%), Positives = 379/861 (43%), Gaps = 98/861 (11%)
Query: 22 QLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDN 81
++SY + L K L A RD ++ R+ R + + + + WL D
Sbjct: 23 KVSYTHNLEKNLAALEKTMKELKAKRDDLERRLKREEARGLQRLSEFQVWLDSVATVEDI 82
Query: 82 VDQLLQMAKSE-KNSCFGH-CPNWIWR-YSVGRKLSKKKRNLKLYIEEGRQYIEIERPAS 138
+ LL+ E + C C + R Y G+ + + R ++ +G + I AS
Sbjct: 83 IITLLRDRNVEIQRLCLCRFCSKSLTRSYRYGKSVFLRLREVEKL--KGEVFGVITEQAS 140
Query: 139 LSAGYFSAERCWE--FDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGK--- 193
SA ER + + ++ L +D V ++G+YGMGG GKT L ++
Sbjct: 141 TSA---FEERPLQPTIVGQDTMLDKAGKHLMEDGVGIMGMYGMGGVGKTTLLTQLYNMFN 197
Query: 194 --RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKR-LCM-RLTQEDR 249
+CG FD ++V +S VE++Q++IA L E + D+S++ +C+ + +E
Sbjct: 198 KDKCG--FDIGIWVVVSQEFHVEKVQDEIAQKLGLGGDEWTQKDKSQKGICLYNILREKS 255
Query: 250 VLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETW 309
++ LDD+W+ +D IG+P KG K+ T+RS+ VC M + +++ L + +
Sbjct: 256 FVLFLDDIWEKVDLAEIGVPDPRTKKGRKLAFTTRSQEVCARMGVEHPMEVQCLEENVAF 315
Query: 310 DLFQKQ-ALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLR 367
DLFQK+ + G+ I +AR ++ +C GLP+A + ++ K + EW+ A+ L
Sbjct: 316 DLFQKKVGQTTLGSDPGIPQLARIVAKKCCGLPLALNVIGETMSCKRTIQEWRHAIHVLN 375
Query: 368 SSKPVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLG 426
S I G+++ L+ SYDNL E+ KS L C+++PED +I E L I
Sbjct: 376 SYAAEFI--GMEDKVLPLLKYSYDNLKGEQVKSSLLYCALYPEDAKILKEDLIEHWICEE 433
Query: 427 IVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEG---KCVKMHDLVRNVAHWIAENEIKC 483
I+ E A ++ L+ + LL++ ++G + V MHD+VR +A WIA +E+
Sbjct: 434 IIDGSEGIEKAEDKGYEIIGCLVRASLLMEWDDGDGRRAVCMHDVVREMALWIA-SELGI 492
Query: 484 ASE----------------KDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQI-------- 519
E K+ + SL S +C L L +
Sbjct: 493 QKEAFIVRAGVGVREIPKIKNWNVVRRMSLMENKIHHLVGSYECMELTTLLLGKREYGSI 552
Query: 520 -HTYTQVSDEIFKGMRMLRVLFL-YNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISF 577
+S E F M L VL L +NK P ++L SL L +L+++ +
Sbjct: 553 RSQLKTISSEFFNCMPKLAVLDLSHNKSLFELPEEISNLVSLKYLN-LLYTEISHLP-KG 610
Query: 578 VGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDL--SECGMERNPFEVIARHTELEELF 635
+ ++KK+ + L +E ++ L NL++L L S + N + + LE L
Sbjct: 611 IQELKKIIHLNLEYTRKLESITGISSLHNLKVLKLFRSRLPWDLNTVKELETLEHLEILT 670
Query: 636 FA-DCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAI 694
D R+K + + S ++L+ Y + S LN H L L S +
Sbjct: 671 TTIDPRAKQFLSSHRLLSHSRLLEIYGSSVSS---------LNRH----LESLSVSTDKL 717
Query: 695 KDLAEKAEVLCIAGIE-GGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLF 753
++ K+ I+ I+ GG N + L ++ I + +G+ L T LI F
Sbjct: 718 REFQIKS--CSISEIKMGGICNFL--------SLVDVNIFNCEGLREL--TFLI-----F 760
Query: 754 FCKLHWLRIEHMKHLGALYNGQMPLSGH------FENLEDLYISHCPKLTRLFTLAVAQN 807
K+ L + H K L + N + G F L L + PKL +++ +
Sbjct: 761 APKIRSLSVWHAKDLEDIINEEKACEGEESGILPFPELNFLTLHDLPKLKKIYWRPLP-- 818
Query: 808 LAQLEKLQVLSCPELQHILID 828
LE++ + CP L+ + +D
Sbjct: 819 FLCLEEINIRECPNLRKLPLD 839
>At1g51480 hypothetical protein
Length = 941
Score = 167 bits (423), Expect = 3e-41
Identities = 205/880 (23%), Positives = 365/880 (41%), Gaps = 133/880 (15%)
Query: 33 VQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQLLQMAKSE 92
+ DL L RD + RV+ + + + V+ W+ I LL+ +E
Sbjct: 122 LDDLHTTMEELKNGRDDLLRRVSIEEDKGLQQLAQVKGWISRVEIVESRFKDLLEDKSTE 181
Query: 93 KNSC--FGHCP-NWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEI-----------ERPAS 138
FG C N I Y+ G K+ K +K + +++ E+ E+
Sbjct: 182 TGRLCLFGFCSENCISSYNYGEKVMKNLEEVKELLS--KKHFEVVAHKIPVPKVEEKNIH 239
Query: 139 LSAG-YFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRCGN 197
+ G Y E W+ +L +D++ + L+GMGG GKT L + +
Sbjct: 240 TTVGLYAMVEMAWK-------------SLMNDEIRTLCLHGMGGVGKTTLLACINNKFVE 286
Query: 198 L---FDQVLFVPISSTVEVERIQEKIAGSLEFEFQ-EKDEMDRSKRLCMRLTQEDRVLVI 253
L FD V++V +S ++E IQ++I G L + + E++ ++ L + + +++
Sbjct: 287 LESEFDVVIWVVVSKDFQLEGIQDQILGRLRLDKEWERETENKKASLINNNLKRKKFVLL 346
Query: 254 LDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQ 313
LDD+W +D + IG+P G KI+ T RS+ V M +I++S L+ DE W+LF+
Sbjct: 347 LDDLWSEVDLNKIGVPPPTRENGAKIVFTKRSKEVSKYMKADMQIKVSCLSPDEAWELFR 406
Query: 314 ---KQALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSS 369
++S I +AR ++ +C GLP+A + + ++ K + EW A++ L S
Sbjct: 407 ITVDDVILSSHE--DIPALARIVAAKCHGLPLALIVIGEAMACKETIQEWHHAINVLNSP 464
Query: 370 KPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVG 429
+ L+ SYD+L E K FL CS+FPED EI E L I G +
Sbjct: 465 AGHKFPGMEERILLVLKFSYDSLKNGEIKLCFLYCSLFPEDFEIEKEKLIEYWICEGYIN 524
Query: 430 EVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENEIKCASEKDI 489
+G N+ L+ + LL++ VKMH ++R +A WI + K +
Sbjct: 525 PNRYEDGGTNQGYDIIGLLVRAHLLIECELTTKVKMHYVIREMALWINSDFGKQQETICV 584
Query: 490 MTLEHTSLRYLWCEKFPNSLDCSNLDFLQIHTYTQVSDEIFKGMRMLRVLFLYNKGRERR 549
+ H + PN ++++ + + +S +I K
Sbjct: 585 KSGAHVRM-------IPN-----DINWEIVRQVSLISTQIEK------------------ 614
Query: 550 PLLTTSLKSLTNLRCILFSKWDLVDIS--FVGDMKKLESITL-CDCSFVELPDVVTQLTN 606
++ S K +NL +L LV+IS F M KL + L + S +ELP+ ++ L +
Sbjct: 615 --ISCSSK-CSNLSTLLLPYNKLVNISVGFFLFMPKLVVLDLSTNMSLIELPEEISNLCS 671
Query: 607 LRLLDLSECGMERNPFEVIARHTELEELFFADCRSKWEVEFLKEFSVP----QVLQRYQI 662
L+ L+LS G++ P +L +L + + +++E L S QVL+ +
Sbjct: 672 LQYLNLSSTGIKSLP----GGMKKLRKLIYLNLEFSYKLESLVGISATLPNLQVLKLFYS 727
Query: 663 QLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKD-------LAEKAEVLCIAGIE----- 710
+ E L H L + + +A I + LA LC+ +
Sbjct: 728 NV--CVDDILMEELQHMDHLKILTVTIDDAMILERIQGIDRLASSIRGLCLTNMSAPRVV 785
Query: 711 ------GGAKNII---PDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLR 761
GG + + ++ + K R+ +E T G ++ ++
Sbjct: 786 LSTTALGGLQQLAILSCNISEIKMDWKSKERREVSPMEIHPSTSTSSPGFKQLSSVNIMK 845
Query: 762 IEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNL------AQLEKLQ 815
+ + L L Q NL+ L++ P++ + ++ +LE L
Sbjct: 846 LVGPRDLSWLLFAQ--------NLKSLHVGFSPEIEEIINKEKGSSITKEIAFGKLESLV 897
Query: 816 VLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVREC 855
+ PEL+ I ++YR L P + F V++C
Sbjct: 898 IYKLPELKEI----------CWNYRTL--PNSRYFDVKDC 925
>At5g43740 disease resistance protein
Length = 862
Score = 167 bits (422), Expect = 4e-41
Identities = 226/903 (25%), Positives = 376/903 (41%), Gaps = 157/903 (17%)
Query: 28 CFNNFVQDLAKEESNLAAI----------RDSVQDRVTRAKKQTRKTAEVVEKWLKDANI 77
CF + + ESNL A+ RD + RV+ + + + V WL I
Sbjct: 19 CFLSDRNYIHMMESNLDALQKTMEELKNGRDDLLGRVSIEEDKGLQRLAQVNGWLSRVQI 78
Query: 78 AMDNVDQLLQMAKSEKNSC--FGHCP-NWIWRYSVGRKLSK---------KKRNLKLYIE 125
LL+ E G+C + I Y+ G K+SK K++ ++ +
Sbjct: 79 VESEFKDLLEAMSIETGRLCLLGYCSEDCISSYNYGEKVSKMLEEVKELLSKKDFRMVAQ 138
Query: 126 EGRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKT 185
E +E + + E W +L +D++ +GLYGMGG GKT
Sbjct: 139 EIIHKVEKKLIQTTVGLDKLVEMAWS-------------SLMNDEIGTLGLYGMGGVGKT 185
Query: 186 MLAMEVGKRCGNL---FDQVLFVPISSTVEVERIQEKIAGSLEFEFQ-EKDEMDRSKRLC 241
L + + L FD V++V +S + E IQ++I G L + + E++ + L
Sbjct: 186 TLLESLNNKFVELESEFDVVIWVVVSKDFQFEGIQDQILGRLRSDKEWERETESKKASLI 245
Query: 242 MRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLS 301
+ + +++LDD+W +D IG+P G KI+ T+RS VC M K+I+++
Sbjct: 246 YNNLERKKFVLLLDDLWSEVDMTKIGVPPPTRENGSKIVFTTRSTEVCKHMKADKQIKVA 305
Query: 302 TLTNDETWDLFQKQALISEGTWI-----SIKNMAREISNECKGLPVATVAVASSLKGKAE 356
L+ DE W+LF+ ++ G I I +AR ++ +C GLP+A + ++ K
Sbjct: 306 CLSPDEAWELFR----LTVGDIILRSHQDIPALARIVAAKCHGLPLALNVIGKAMSCKET 361
Query: 357 V-EWKVALDRLRSSK---PVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCE 412
+ EW A++ L S+ P E+ L L+ SYD+L E K FL CS+FPED E
Sbjct: 362 IQEWSHAINVLNSAGHEFPGMEERIL----PILKFSYDSLKNGEIKLCFLYCSLFPEDSE 417
Query: 413 IPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNV 472
IP E I G + +G N L+ + LL++ VKMHD++R +
Sbjct: 418 IPKEKWIEYWICEGFINPNRYEDGGTNHGYDIIGLLVRAHLLIECELTDNVKMHDVIREM 477
Query: 473 AHWI-----AENEIKCASE--------KDI-------MTLEHTSLRYLWC-EKFPNSLDC 511
A WI + E C DI M+ T ++ + C K PN
Sbjct: 478 ALWINSDFGKQQETICVKSGAHVRMIPNDINWEIVRTMSFTCTQIKKISCRSKCPN---L 534
Query: 512 SNLDFLQIHTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWD 571
S L L ++S+ F+ M L VL L NL I +
Sbjct: 535 STLLILDNRLLVKISNRFFRFMPKLVVLDL-----------------SANLDLIKLPE-- 575
Query: 572 LVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTEL 631
+IS +G ++ L +I+L LP + +L L L+L G+ + + A L
Sbjct: 576 --EISNLGSLQYL-NISL--TGIKSLPVGLKKLRKLIYLNLEFTGVHGSLVGIAATLPNL 630
Query: 632 EEL-FFADC--------RSKWEVEFLKEFSV----PQVLQRYQIQLGSMFSGFQDEFLNH 678
+ L FF C + ++E LK + +L+R Q D +
Sbjct: 631 QVLKFFYSCVYVDDILMKELQDLEHLKILTANVKDVTILERIQ---------GDDRLASS 681
Query: 679 HRTLFLSYLDTSNAAIKDLAEKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIR-DSKG 737
R+L L + T + +A + G++ + M ++ E+ I +SK
Sbjct: 682 IRSLCLEDMSTPRVILSTIA-------LGGLQ--------QLAILMCNISEIRIDWESKE 726
Query: 738 IECLVDT-CLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSG--HFENLEDLYISHCP 794
L T L G+ F +L + I ++ GQ LS + +NL+ L + P
Sbjct: 727 RRELSPTEILPSTGSPGFKQLSTVYINQLE-------GQRDLSWLLYAQNLKKLEVCWSP 779
Query: 795 KLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDD--DRDEISAYDYRLLLFPKLKKFHV 852
++ + N+ +L + V+ L+ + + D EI ++YR L P L+K ++
Sbjct: 780 QIEEIINKEKGMNITKLHRDIVVPFGNLEDLALRQMADLTEI-CWNYRTL--PNLRKSYI 836
Query: 853 REC 855
+C
Sbjct: 837 NDC 839
>At5g05400 NBS/LRR disease resistance protein
Length = 874
Score = 165 bits (417), Expect = 1e-40
Identities = 161/634 (25%), Positives = 276/634 (43%), Gaps = 84/634 (13%)
Query: 29 FNNFVQDLA---KEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQL 85
F N V +A K L A RD + R+ + + + V++WL + + +
Sbjct: 29 FRNLVDHVAALKKTVRQLEARRDDLLKRIKVQEDRGLNLLDEVQQWLSEVESRVCEAHDI 88
Query: 86 LQMAKSE-KNSCFG-HCPNWI-WRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLSAG 142
L + E N C G +C + Y + + K ++++ + +G + E+ + +
Sbjct: 89 LSQSDEEIDNLCCGQYCSKRCKYSYDYSKSVINKLQDVENLLSKG-VFDEVAQKGPIPK- 146
Query: 143 YFSAERCW--EFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKR---CGN 197
ER + E ++ E ++ + V ++G+YGMGG GKT L ++ + N
Sbjct: 147 --VEERLFHQEIVGQEAIVESTWNSMMEVGVGLLGIYGMGGVGKTTLLSQINNKFRTVSN 204
Query: 198 LFDQVLFVPISSTVEVERIQEKIAGSLEFE---FQEKDEMDRSKRLCMRLTQEDRVLVIL 254
FD ++V +S V+RIQE I L+ +++K E + + + R + + +++L
Sbjct: 205 DFDIAIWVVVSKNPTVKRIQEDIGKRLDLYNEGWEQKTENEIASTI-KRSLENKKYMLLL 263
Query: 255 DDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQK 314
DD+W +D IGIP + G KI TSRS VC M K+I+++ L D+ WDLF +
Sbjct: 264 DDMWTKVDLANIGIP-VPKRNGSKIAFTSRSNEVCGKMGVDKEIEVTCLMWDDAWDLFTR 322
Query: 315 QALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVE-WKVALDRLRSSKPVN 373
+ + I +A+ I+ +C GLP+A + ++ K +E W A V
Sbjct: 323 NMKETLESHPKIPEVAKSIARKCNGLPLALNVIGETMARKKSIEEWHDA---------VG 373
Query: 374 IEKGLQ-NPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVH 432
+ G++ + L+ SYD+L E+ KS FL ++FPED EI + L +G GI+
Sbjct: 374 VFSGIEADILSILKFSYDDLKCEKTKSCFLFSALFPEDYEIGKDDLIEYWVGQGII---L 430
Query: 433 SYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENEIKCASEK----- 487
+G + L + LL + + VKMHD+VR +A WI+ C +K
Sbjct: 431 GSKGINYKGYTIIGTLTRAYLLKESETKEKVKMHDVVREMALWISSG---CGDQKQKNVL 487
Query: 488 ---------------DIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQI--HTYTQVSDEIF 530
D + SL Y E+ SL C L+ L + + ++S E
Sbjct: 488 VVEANAQLRDIPKIEDQKAVRRMSLIYNQIEEACESLHCPKLETLLLRDNRLRKISREFL 547
Query: 531 KGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLC 590
+ +L VL L SL +L+++ + L + L
Sbjct: 548 SHVPILMVLDL-------------SLNP------------NLIELPSFSPLYSLRFLNLS 582
Query: 591 DCSFVELPDVVTQLTNLRLLDLSECGMERNPFEV 624
LPD + L NL L+L M + +E+
Sbjct: 583 CTGITSLPDGLYALRNLLYLNLEHTYMLKRIYEI 616
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,310,953
Number of Sequences: 26719
Number of extensions: 1048517
Number of successful extensions: 4089
Number of sequences better than 10.0: 208
Number of HSP's better than 10.0 without gapping: 133
Number of HSP's successfully gapped in prelim test: 75
Number of HSP's that attempted gapping in prelim test: 3011
Number of HSP's gapped (non-prelim): 697
length of query: 1094
length of database: 11,318,596
effective HSP length: 110
effective length of query: 984
effective length of database: 8,379,506
effective search space: 8245433904
effective search space used: 8245433904
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC137995.11


