

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137838.4 + phase: 0
(496 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
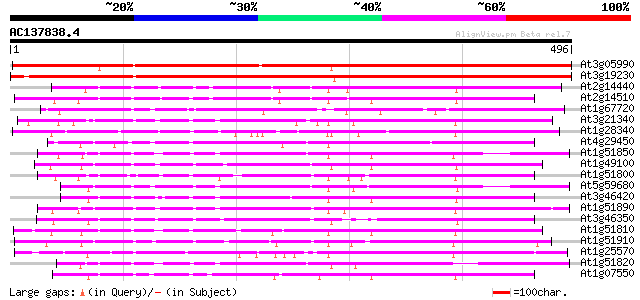
At3g05990 unknown protein 597 e-171
At3g19230 hypothetical protein 498 e-141
At2g14440 putative receptor-like protein kinase 215 5e-56
At2g14510 putative receptor-like protein kinase 200 1e-51
At1g67720 putative receptor protein kinase 189 3e-48
At3g21340 serine/threonine-specific protein kinase, putative 178 7e-45
At1g28340 unknown protein 177 2e-44
At4g29450 serine/threonine-specific receptor protein kinase-like... 174 1e-43
At1g51850 Putative protein kinase 172 3e-43
At1g49100 putative receptor-like protein kinase gb|AAD15465.1 172 5e-43
At1g51800 unknown protein 171 7e-43
At5g59680 serine/threonine-specific protein kinase - like 169 3e-42
At3g46420 hypothetical protein 168 7e-42
At1g51890 Putative protein kinase 166 3e-41
At3g46350 receptor-like protein kinase homolog 165 6e-41
At1g51810 Putative protein kinase 164 1e-40
At1g51910 Putative protein kinase 163 2e-40
At1g25570 hypothetical protein 163 2e-40
At1g51820 Putative protein kinase 161 7e-40
At1g07550 protein kinase like 161 7e-40
>At3g05990 unknown protein
Length = 517
Score = 597 bits (1540), Expect = e-171
Identities = 292/504 (57%), Positives = 376/504 (73%), Gaps = 12/504 (2%)
Query: 3 ILFFLLLFISFHTPSFSQ-TPPKGFLINCGTLTTTQINNRTWLPDSNFITTGTPKNITTQ 61
+L LL S + S SQ +PP LI+CG +++ I+ R W PD F+++GTPKN++ Q
Sbjct: 4 LLLLLLTVFSLLSSSLSQPSPPSAILIDCGASSSSVIDGRQWQPDETFVSSGTPKNVSDQ 63
Query: 62 VLLPTLKTLRSFPLQVK----KHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQI 117
VL L T+RSFPL + K CY + V RG KYMIRTTY+YGGVNG P PPVFDQI
Sbjct: 64 VLDEILFTVRSFPLSLDGTHHKFCYVMSVSRGWKYMIRTTYYYGGVNGKGTP-PPVFDQI 122
Query: 118 IDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGD 177
+DGTLW +VNTT DYA+G +S+YEGVFLA GK +S C+ SNSYT SDPF+SALE + L
Sbjct: 123 VDGTLWGIVNTTADYADGLASYYEGVFLAQGKSISVCVASNSYTTSDPFISALELVRLDG 182
Query: 178 SLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTENVSVSGF 237
+LYN+TDF + LVAR++FGYSGP IR+PDDQFDR WEP+ NST N + VSGF
Sbjct: 183 TLYNSTDFTTVGMSLVARHAFGYSGPIIRFPDDQFDRFWEPYSL-NSTVPNNRKLEVSGF 241
Query: 238 WNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADNTA----GSRIFNISVN 293
WNLPPS++F T L + Q++ LE WP L + YYIALYFA ++ GSR+F++SVN
Sbjct: 242 WNLPPSRIFNTDLRATQVQPLEFTWPPMPLKMATYYIALYFAHDSDSMGDGSRVFDVSVN 301
Query: 294 GVHYYRDLNAIASGVVVFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVLSLGGRTS 353
G+ YY++L+ +G V+FA++WPL G TT+ L+P + S+L PLIN GE+F +LSLGG+T
Sbjct: 302 GITYYKELSVTPAGAVIFASRWPLEGLTTLALSPRSGSNLPPLINGGEMFELLSLGGKTL 361
Query: 354 TRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSL 413
RDV AL +K S +N P DWSGDPC+P+ YSW+GI+CSEG RIR+V LNLT+M +SGSL
Sbjct: 362 VRDVTALNAIKNSFKNAPADWSGDPCLPKNYSWSGISCSEGPRIRVVALNLTNMGVSGSL 421
Query: 414 SSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEV 473
+ VA +TAL++IWLGNNSLSG +P+ SSL LE+LH E+N FSG IPSSLG + L+E+
Sbjct: 422 APAVAKLTALSSIWLGNNSLSGSLPDFSSLKRLESLHFEDNLFSGSIPSSLGGVPHLREL 481
Query: 474 FLQNNNLTGQIPANLL-KPGLSIR 496
FLQNNNLTGQ+P+NLL KPGL++R
Sbjct: 482 FLQNNNLTGQVPSNLLQKPGLNLR 505
>At3g19230 hypothetical protein
Length = 519
Score = 498 bits (1282), Expect = e-141
Identities = 245/504 (48%), Positives = 346/504 (68%), Gaps = 13/504 (2%)
Query: 1 MSILFFLLLFISFHTPSFSQTPPKGFLINCGTLTTTQINNRTWLPDSNFITTGTPKNITT 60
MS+ FFLLL S S + PP+GF +NCG+ ++T +N + PD FI+ G I
Sbjct: 1 MSLFFFLLLCFSI---SHALPPPRGFFLNCGSSSSTNLNEIEYTPDEGFISVGNTTTIKQ 57
Query: 61 QVLLPTLKTLRSFP-LQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIID 119
+ L+P L TLR FP +KHCYN PV + +KY+IRTTY+YG +G ++P PVFDQII
Sbjct: 58 KDLVPILSTLRYFPDKSSRKHCYNFPVAKTSKYLIRTTYYYGNFDGKNNP--PVFDQIIG 115
Query: 120 GTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYT-DSDPFVSALEFLILGDS 178
GT WSVVNT+ DYA G SS+YE + G +S C+ N++T S PF+S+L+ L D+
Sbjct: 116 GTKWSVVNTSEDYAKGQSSYYEIIVGVPGNRLSVCLAKNAHTLSSSPFISSLDVQSLEDT 175
Query: 179 LYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFG-QSNSTKANTENVSVSGF 237
+YN+TD ++ + L+ARNSFG G I YPDD+++R+W+PF Q + T + ++ S F
Sbjct: 176 MYNSTDLGSYKLSLIARNSFGGDGEIISYPDDKYNRLWQPFSDQKHLTVTSRSRINPSNF 235
Query: 238 WNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFADNTAGS----RIFNISVN 293
WN+PP++ F + + + LEL+WP LP++KYY+ALYF DN + R F++SVN
Sbjct: 236 WNIPPAEAFVEGFTASKGKPLELQWPPFPLPATKYYVALYFQDNRSPGPMSWRAFDVSVN 295
Query: 294 GVHYYRDLNAIASGVVVFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVLSLGGRTS 353
G+ + R LN +GV+V++ QWPLSG T ITLTP+ + +GP INAGEVF +L LGG T+
Sbjct: 296 GLSFLRKLNVSTNGVMVYSGQWPLSGQTQITLTPAKDAPVGPFINAGEVFQILPLGGTTN 355
Query: 354 TRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDLSGSL 413
+D IA++ + ES+ PP+DWSGDPC+PR SWTG+TCS+ R+++LNLT++ LSGSL
Sbjct: 356 IKDAIAMEDLLESIMKPPVDWSGDPCLPRANSWTGLTCSKDKIARVISLNLTNLGLSGSL 415
Query: 414 SSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEV 473
+ MTAL ++WLG N L+G IP+LS +T LETLHLE+NQF+G IP SL + SL+ +
Sbjct: 416 PPSINKMTALKDLWLGKNKLTGPIPDLSPMTRLETLHLEDNQFTGAIPESLAKLPSLRTL 475
Query: 474 FLQNNNLTGQIPANLL-KPGLSIR 496
++NN L G IP+ LL + GL+I+
Sbjct: 476 SIKNNKLKGTIPSVLLQRKGLTIQ 499
>At2g14440 putative receptor-like protein kinase
Length = 886
Score = 215 bits (547), Expect = 5e-56
Identities = 151/470 (32%), Positives = 239/470 (50%), Gaps = 31/470 (6%)
Query: 38 INNRTWLPDSNFITTGTPKNITTQVLLP----TLKTLRSFPLQVKKHCYNIPVYRGAKYM 93
+ N T++ D NF+ G NI + K LR FP ++ +CY++ V +G KY+
Sbjct: 47 LTNLTYISDVNFVRGGKTGNIKNNSDIDFTSRPYKVLRYFPEGIR-NCYSLSVKQGTKYL 105
Query: 94 IRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTT-VDYANGNSSFYEGVFLAVGKFMS 152
IRT +FYG +G++ T P FD + +W+ V+ VD G+ E + + +
Sbjct: 106 IRTLFFYGNYDGLN--TSPRFDLFLGPNIWTSVDVQKVD--GGDGVIEEIIHVTRCNILD 161
Query: 153 FCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQF 212
C+ T P +SA+E L Y+T ++ + F SG +RYP+D +
Sbjct: 162 ICLVKTGTTT--PMISAIELRPLR---YDTYTARTGSLKKILHFYFTNSGKEVRYPEDVY 216
Query: 213 DRIWEPFGQSNSTKANTENVSVSGF---WNLPPSKVFETHLGSEQLESLELRWPTASLPS 269
DR+W P Q T+ NT +VSGF +N P + + + E L W + S
Sbjct: 217 DRVWIPHSQPEWTQINTTR-NVSGFSDGYNPPQDVIKTASIPTNVSEPLTFTWMSES-SD 274
Query: 270 SKYYIALYFAD----NTAGSRIFNISVNGVHY--YRDLNAIASGVVVFANQWPLSGPTTI 323
+ Y LYFA+ +R F I VNGV+Y Y A ++ A G +
Sbjct: 275 DETYAYLYFAEIQQLKANETRQFKILVNGVYYIDYIPRKFEAETLITPAALKCGGGVCRV 334
Query: 324 TLTPSASSSLGPLINAGEVFNVLSLG-GRTSTRDVIALQRVKESLRNPPLDWSGDPCVPR 382
L+ + S+L P +NA E+F+V+ T+T +VIA++ ++ + + + W GDPCVP
Sbjct: 335 QLSKTPKSTLPPQMNAIEIFSVIQFPQSDTNTDEVIAIKNIQSTYKVSRISWQGDPCVPI 394
Query: 383 QYSWTGITCSE---GLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP- 438
Q+SW G++C+ RI++L+L+S L+G ++ + N+T L + L NN+L+G IP
Sbjct: 395 QFSWMGVSCNVIDISTPPRIISLDLSSSGLTGVITPSIQNLTMLRELDLSNNNLTGVIPP 454
Query: 439 NLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANL 488
+L +LTML L L N +GE+P L I L + L+ NNL G +P L
Sbjct: 455 SLQNLTMLRELDLSNNNLTGEVPEFLATIKPLLVIHLRGNNLRGSVPQAL 504
>At2g14510 putative receptor-like protein kinase
Length = 868
Score = 200 bits (509), Expect = 1e-51
Identities = 147/485 (30%), Positives = 244/485 (50%), Gaps = 37/485 (7%)
Query: 5 FFLLLFISFHTPSFSQTP-PKGFL-INCGTLTTTQI----NNRTWLPDSNFITTGTPKNI 58
F LL +F S ++ +GF+ ++CG + +N T++ D NFI G NI
Sbjct: 7 FMLLACATFSIMSLVKSQNQQGFISLDCGLPSKESYIEPSSNLTFISDVNFIRGGKTGNI 66
Query: 59 T----TQVLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVF 114
T + K LR FP ++ +CY++ V +G KY+IRT ++YG +G++ T P F
Sbjct: 67 QNNSRTNFIFKPFKVLRYFPDGIR-NCYSLSVKQGTKYLIRTLFYYGNYDGLN--TSPRF 123
Query: 115 DQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLI 174
D + +W+ V+ + G+ E V + + C+ S P +SA+E
Sbjct: 124 DLFLGPNIWTSVDVLIADV-GDGVVEEIVHVTRSNILDICLVKTG--TSTPMISAIELRP 180
Query: 175 LGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTENVSV 234
L Y+T ++ +A F S +IRYP+D +DR+W P+ Q T+ NT +V
Sbjct: 181 LR---YDTYTARTGSLKSMAHFYFTNSDEAIRYPEDVYDRVWMPYSQPEWTQINTTR-NV 236
Query: 235 SGF---WNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFAD----NTAGSRI 287
SGF +N P + + + E L W S + Y L+FA+ +R
Sbjct: 237 SGFSDGYNPPQGVIQTASIPTNGSEPLTFTWNLES-SDDETYAYLFFAEIQQLKVNETRE 295
Query: 288 FNISVNGVHYYRDLNAIASGVVVFANQWPLS---GPTTITLTPSASSSLGPLINAGEVFN 344
F I NGV Y D +N PL G + L+ + S+L PL+NA E+F+
Sbjct: 296 FKILANGVDYI-DYTPWKFEARTLSNPAPLKCEGGVCRVQLSKTPKSTLPPLMNAIEIFS 354
Query: 345 VLSLG-GRTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCSE---GLRIRIV 400
V+ T+T +VIA+++++ + + + W GDPCVP+Q+SW G++C+ RI+
Sbjct: 355 VIQFPQSDTNTDEVIAIKKIQSTYQLSRISWQGDPCVPKQFSWMGVSCNVIDISTPPRII 414
Query: 401 TLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGE 459
+L+L+ L+G +S + N+T L + L NN+L+G++P L+++ L +HL N G
Sbjct: 415 SLDLSLSGLTGVISPSIQNLTMLRELDLSNNNLTGEVPEFLATIKPLLVIHLRGNNLRGS 474
Query: 460 IPSSL 464
+P +L
Sbjct: 475 VPQAL 479
>At1g67720 putative receptor protein kinase
Length = 914
Score = 189 bits (480), Expect = 3e-48
Identities = 148/501 (29%), Positives = 237/501 (46%), Gaps = 60/501 (11%)
Query: 28 INCGTLTTTQINNRT---WLPDSNFITTGTPKNIT-TQVLLPTLKTLRSFPLQVKKHCYN 83
I+CG ++ + RT W+ DS I G P + T + R FP KK+CY
Sbjct: 29 IDCGC-SSNYTDPRTGLGWVSDSEIIKQGKPVTLANTNWNSMQYRRRRDFPTDNKKYCYR 87
Query: 84 IPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYANGNSSFYEGV 143
+ +Y++RTT+ YGG+ + P F +D T W+ TV + + E +
Sbjct: 88 LSTKERRRYIVRTTFLYGGLGSEE--AYPKFQLYLDATKWA----TVTIQEVSRVYVEEL 141
Query: 144 FL-AVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSG 202
+ A ++ C+ + T S PF+S LE L S+Y T +NF + + AR +FG
Sbjct: 142 IVRATSSYVDVCVCC-AITGS-PFMSTLELRPLNLSMYATDYEDNFFLKVAARVNFGAPN 199
Query: 203 -PSIRYPDDQFDRIWEPFGQSN----------STKANTENVSVSGFWNLPPSKVFETHL- 250
++RYPDD +DRIWE +T+ NT + PP KV +T +
Sbjct: 200 MDALRYPDDPYDRIWESDINKRPNYLVGVAPGTTRINTSKTINTLTREYPPMKVMQTAVV 259
Query: 251 GSEQLESLELRWPTASLPSSKYYIALYFADNTAGSRIFNISVNGVH-------YYRDLNA 303
G++ L S L + Y YFA+ I + N Y+ D +
Sbjct: 260 GTQGLISYRLNLEDFPANARAY---AYFAE------IEELGANETRKFKLVQPYFPDYSN 310
Query: 304 IASGVVVFAN-QWPLSGPTTITLT----------PSASSSLGPLINAGEVFNVLSLGGRT 352
+ AN + L P+ + +T + S+ GPL+NA E+ L + +T
Sbjct: 311 AVVNIAENANGSYTLYEPSYMNVTLDFVLTFSFGKTKDSTQGPLLNAIEISKYLPISVKT 370
Query: 353 STRDVIALQRVKESLRNPPLDWS---GDPCVPRQYSWTGITCSEGLRIRIVTLNLTSMDL 409
DV L ++ +P DW+ GDPC+P +SW + CS R+ + L+ +L
Sbjct: 371 DRSDVSVLDAIRSM--SPDSDWASEGGDPCIPVLWSW--VNCSSTSPPRVTKIALSRKNL 426
Query: 410 SGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISS 469
G + + M ALT +WL +N L+G +P++S L L+ +HLE NQ SG +P L ++ +
Sbjct: 427 RGEIPPGINYMEALTELWLDDNELTGTLPDMSKLVNLKIMHLENNQLSGSLPPYLAHLPN 486
Query: 470 LKEVFLQNNNLTGQIPANLLK 490
L+E+ ++NN+ G+IP+ LLK
Sbjct: 487 LQELSIENNSFKGKIPSALLK 507
>At3g21340 serine/threonine-specific protein kinase, putative
Length = 880
Score = 178 bits (451), Expect = 7e-45
Identities = 146/501 (29%), Positives = 240/501 (47%), Gaps = 43/501 (8%)
Query: 8 LLFISFHT--PSFSQTPPKGFL-INCGTLTTTQINNR-----TWLPDSNFITTGTP---- 55
L+FISF+ KGF+ ++CG+L N T+ D F+ +G
Sbjct: 13 LIFISFYALLHLVEAQDQKGFISLDCGSLPNEPPYNDPSTGLTYSTDDGFVQSGKTGRIQ 72
Query: 56 KNITTQVLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFD 115
K + P+LK LR FP ++CY + V + Y+I+ + YG +G+++P P FD
Sbjct: 73 KAFESIFSKPSLK-LRYFP-DGFRNCYTLNVTQDTNYLIKAVFVYGNYDGLNNP--PSFD 128
Query: 116 QIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLIL 175
+ LW V+ N + E + + K + C+ S P ++ LE L
Sbjct: 129 LYLGPNLWVTVDMN---GRTNGTIQEIIHKTISKSLQVCLVKTG--TSSPMINTLELRPL 183
Query: 176 GDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTENVSVS 235
++ YNT + ++ R F SG +IRYPDD DR W PF + T N++++
Sbjct: 184 KNNTYNT---QSGSLKYFFRYYFSGSGQNIRYPDDVNDRKWYPFFDAKEWTELTTNLNIN 240
Query: 236 GFWNLPPSKVFETHLGS--EQLESLELRWPTASLPSS--KYYIALYFAD----NTAGSRI 287
P +V + + W LPSS ++Y+ ++FA+ + +R
Sbjct: 241 SSNGYAPPEVVMASASTPISTFGTWNFSW---LLPSSTTQFYVYMHFAEIQTLRSLDTRE 297
Query: 288 FNISVNGVHYYRDLN--AIASGVVVFAN-QWPLSGPTTITLTPSASSSLGPLINAGEVFN 344
F +++NG Y + +A+ + ++ Q G + LT + S+L PL+NA EVF
Sbjct: 298 FKVTLNGKLAYERYSPKTLATETIFYSTPQQCEDGTCLLELTKTPKSTLPPLMNALEVFT 357
Query: 345 VLSLGG-RTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCS--EGLRIRIVT 401
V+ T+ DV A++ ++ + + W GDPCVP+Q+ W G+ C+ + IVT
Sbjct: 358 VIDFPQMETNPDDVAAIKSIQSTYGLSKISWQGDPCVPKQFLWEGLNCNNLDNSTPPIVT 417
Query: 402 -LNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGE 459
LNL+S L+G ++ + N+T L + L NN+L+G IP L+ + L ++L N F+G
Sbjct: 418 SLNLSSSHLTGIIAQGIQNLTHLQELDLSNNNLTGGIPEFLADIKSLLVINLSGNNFNGS 477
Query: 460 IPSSLGNISSLKEVFLQNNNL 480
IP L LK + N NL
Sbjct: 478 IPQILLQKKGLKLILEGNANL 498
>At1g28340 unknown protein
Length = 697
Score = 177 bits (448), Expect = 2e-44
Identities = 160/519 (30%), Positives = 250/519 (47%), Gaps = 48/519 (9%)
Query: 3 ILFFLLLFISFHTPSFSQTPPKGFLINCGTLTT--TQINNRTWLPDSNFITTGTPKNITT 60
IL LLL S ++ P I+CG T W D + T G P N TT
Sbjct: 6 ILASLLLSSFSLYSSLARPAPYALRISCGARKNVRTPPTYALWFKDIAY-TGGVPANATT 64
Query: 61 QVLL-PTLKTLRSFPL-QVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQII 118
+ P LKTLR FP+ + +CYNI Y +R F+G V+ P+FD I
Sbjct: 65 PTYITPPLKTLRYFPISEGPNNCYNIVRVPKGHYSVRI--FFGLVDQPSFDKEPLFDISI 122
Query: 119 DGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDS 178
+GT S + + + + F E + +G + C S + DP + ++E L + D
Sbjct: 123 EGTQISSLKSGWS-SQDDQVFAEALIFLLGGTATICFHSTGH--GDPAILSIEILQVDDK 179
Query: 179 LYNTTDFNNFAIGLVARNSF------GYSGPSIRYPDDQF--DRIWE---PFGQS----N 223
Y+ + + G++ R + G S Y D + DR W FG+S
Sbjct: 180 AYSFGE--GWGQGVILRTATRLTCGTGKSRFDEDYRGDHWGGDRFWNRMRSFGKSADSPR 237
Query: 224 STKANTENVSVSGFWNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFA--DN 281
ST+ + SVS N P ++++ L S + +L + P+ Y + L+FA DN
Sbjct: 238 STEETIKKASVSP--NFYPEGLYQSALVSTD-DQPDLTYSLDVEPNRNYSVWLHFAEIDN 294
Query: 282 TA---GSRIFNISVNGVHYYRDLNAIASG-----VVVFANQWPLSGPT-TITLTPSASSS 332
T G R+F++ +NG ++ D++ I +V +SG T T+ L P A
Sbjct: 295 TITAEGKRVFDVVINGDTFFEDVDIIKMSGGRYAALVLNATVTVSGRTLTVVLQPKAGGH 354
Query: 333 LGPLINAGEVFNVLSLGGRTSTRDVIALQRVKESLRNPP-LDWSGDPCVPRQYSWTGITC 391
+INA EVF +++ +T +V ALQ++K++L P W+GDPCVP Q+ W+G C
Sbjct: 355 A--IINAIEVFEIITAEFKTLRDEVSALQKMKKALGLPSRFGWNGDPCVPPQHPWSGANC 412
Query: 392 S---EGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIP-NLSSLTMLE 447
R I L+L + L G L + ++ + L +I L N++ G IP +L S+T LE
Sbjct: 413 QLDKNTSRWFIDGLDLDNQGLKGFLPNDISKLKHLQSINLSENNIRGGIPASLGSVTSLE 472
Query: 448 TLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPA 486
L L N F+G IP +LG ++SL+ + L N+L+G++PA
Sbjct: 473 VLDLSYNSFNGSIPETLGELTSLRILNLNGNSLSGKVPA 511
>At4g29450 serine/threonine-specific receptor protein kinase-like
protein
Length = 863
Score = 174 bits (441), Expect = 1e-43
Identities = 139/453 (30%), Positives = 221/453 (48%), Gaps = 36/453 (7%)
Query: 34 TTTQINNRTWLPDSNFITTGTPKNITTQ-------VLLPTLKTLRSFPLQVKKHCYNIPV 86
T T IN ++ D FITTG ++ + VLL TL +R+FP Q ++CY + +
Sbjct: 43 TMTNIN---YVSDEAFITTGVNFKVSEEYGYPKNPVLLSTLAEVRAFP-QGNRNCYTLKL 98
Query: 87 YRGAK--YMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYANGNSSFYEGVF 144
+G Y+IR ++ YG +G P FD ++ WS TV + N + + +
Sbjct: 99 SQGKDHLYLIRASFMYGNYDG--KKALPEFDLYVNVNFWS----TVKFKNASDQVTKEIL 152
Query: 145 -LAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGP 203
A + C+ + PF+S LE + S+Y T N ++ L R GY
Sbjct: 153 SFAESDTIYVCLVNKG--KGTPFISGLELRPVNSSIYGTEFGRNVSLVLYRRWDIGYLNG 210
Query: 204 SIRYPDDQFDRIWEPFGQSNSTKANTENVSVSGFWN--LPPSKVFETHLGSEQLES-LEL 260
+ RY DD+FDRIW P+ + S + + + F N PP +V +T E ++ LEL
Sbjct: 211 TGRYQDDRFDRIWSPYSSNISWNSIITSGYIDVFQNGYCPPDEVIKTAAAPENVDDPLEL 270
Query: 261 RWPTASLPSSKYYIALYFAD----NTAGSRIFNISVNGVHYYR-DLNAIASGVVVFANQW 315
W T+ P+ ++Y LYFA+ +R I NG + F+N
Sbjct: 271 FW-TSDDPNVRFYAYLYFAELETLEKNETRKIKILWNGSPVSETSFEPSSKYSTTFSNPR 329
Query: 316 PLSGPT-TITLTPSASSSLGPLINAGEVFNVLSLGG-RTSTRDVIALQRVKESLRNPPLD 373
+G I++ + S+L P++NA E+F SL T+ D+ A++ +K + + +
Sbjct: 330 AFTGKDHWISIQKTVDSTLPPILNAIEIFTAQSLDEFSTTIEDIHAIESIKATYKVNKV- 388
Query: 374 WSGDPCVPRQYSWTGITCSE-GLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNS 432
WSGDPC PR + W G+ CS+ +I +LNL+S L G + N++ L ++ L NN
Sbjct: 389 WSGDPCSPRLFPWEGVGCSDNNNNHQIKSLNLSSSGLLGPIVLAFRNLSLLESLDLSNND 448
Query: 433 LSGQIPN-LSSLTMLETLHLEENQFSGEIPSSL 464
L +P L+ L L+ L+L+ N F+G IP SL
Sbjct: 449 LQQNVPEFLADLKHLKVLNLKGNNFTGFIPKSL 481
>At1g51850 Putative protein kinase
Length = 875
Score = 172 bits (437), Expect = 3e-43
Identities = 138/494 (27%), Positives = 233/494 (46%), Gaps = 63/494 (12%)
Query: 25 GFL-INCGTLTTTQINNR-----TWLPDSNFITTGTPKNITTQVL----LPTLKTLRSFP 74
GF+ ++CG N T+ D + G P I + PTL TLR FP
Sbjct: 25 GFISVDCGLAPRESPYNEAKTGLTYTSDDGLVNVGKPGRIAKEFEPLADKPTL-TLRYFP 83
Query: 75 LQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYAN 134
V+ +CYN+ V Y+I+ T+ YG +G++ P FD LW+ V++
Sbjct: 84 EGVR-NCYNLNVTSDTNYLIKATFVYGNYDGLN--VGPNFDLYFGPNLWTTVSS------ 134
Query: 135 GNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVA 194
N + E + + + C+ S PF++ LE + ++Y T ++ +
Sbjct: 135 -NDTIKEIIHVTKTNSLQVCLIKTGI--SIPFINVLELRPMKKNMYVT---QGESLNYLF 188
Query: 195 RNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTE-NVSVSGFWNLPPSKVFETHLGSE 253
R S IR+PDD +DR W P+ ++ T+ T +V+ S + LP S + + +
Sbjct: 189 RVYISNSSTRIRFPDDVYDRKWYPYFDNSWTQVTTTLDVNTSLTYELPQSVMAKAATPIK 248
Query: 254 QLESLELRWPTASLPSSKYYIALYFAD----NTAGSRIFNISVNGVHYYRDLNAIASGVV 309
++L + W T P++K+Y ++FA+ +R FN+++NG++ Y +
Sbjct: 249 ANDTLNITW-TVEPPTTKFYSYMHFAELQTLRANDAREFNVTMNGIYTYGPYSPKPLKTE 307
Query: 310 VFANQWPLS---GPTTITLTPSASSSLGPLINAGEVFNVLSLGG-RTSTRDVIALQRVKE 365
++ P G + + + S+L PL+NA E F V+ T+ DV A++ V++
Sbjct: 308 TIYDKIPEQCDGGACLLQVVKTLKSTLPPLLNAIEAFTVIDFPQMETNGDDVDAIKNVQD 367
Query: 366 SLRNPPLDWSGDPCVPRQYSWTGITC--SEGLRIRIVT-LNLTSMDLSGSLSSFVANMTA 422
+ + W GDPCVP+ + W G+ C S+ I+T L+L+S L+GS++ + N
Sbjct: 368 TYGISRISWQGDPCVPKLFLWDGLNCNNSDNSTSPIITSLDLSSSGLTGSITQAIQN--- 424
Query: 423 LTNIWLGNNSLSGQIPNLSSLTMLETLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTG 482
LT L+ L L +N +GEIP LG+I SL + L NNL+G
Sbjct: 425 --------------------LTNLQELDLSDNNLTGEIPDFLGDIKSLLVINLSGNNLSG 464
Query: 483 QIPANLL-KPGLSI 495
+P +LL K G+ +
Sbjct: 465 SVPPSLLQKKGMKL 478
>At1g49100 putative receptor-like protein kinase gb|AAD15465.1
Length = 888
Score = 172 bits (435), Expect = 5e-43
Identities = 138/473 (29%), Positives = 218/473 (45%), Gaps = 35/473 (7%)
Query: 23 PKGFL-INCGTLT-----TTQINNRTWLPDSNFITTGTPKNITTQV---LLPTLKTLRSF 73
P GF+ ++CG L T T+ DS+FI +G ++ TLR F
Sbjct: 27 PNGFITLDCGLLPDGSPYTNPSTGLTFTSDSSFIESGKNGRVSKDSERNFEKAFVTLRYF 86
Query: 74 PLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYA 133
P +++CYN+ V +G Y+IR + YG +G++ T P FD I + VN A
Sbjct: 87 P-DGERNCYNLNVTQGTNYLIRAAFLYGNYDGLN--TVPNFDLFIGPNKVTTVNFN---A 140
Query: 134 NGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLV 193
G F E + ++ + C+ T P +S LE L Y + ++ L
Sbjct: 141 TGGGVFVEIIHMSRSTPLDICLVKTGTTT--PMISTLELRPLRSDTYISAIGSSLL--LY 196
Query: 194 ARNSFGYSGPSIRYPDDQFDRIWEPFGQSN-STKANTENVSVSGFWNLPPSKVFETHLGS 252
R SG +RYPDD DR W PF T NV+ S ++LP +
Sbjct: 197 FRGYLNDSGVVLRYPDDVNDRRWFPFSYKEWKIVTTTLNVNTSNGFDLPQGAMASAATRV 256
Query: 253 EQLESLELRWPTASLPSSKYYIALYFADNTA----GSRIFNISVNGVHYYRDLNAIASGV 308
+ E W +++++I L+FA+ +R FN+ +NG YY + +
Sbjct: 257 NDNGTWEFPWSLED-STTRFHIYLHFAELQTLLANETREFNVLLNGKVYYGPYSPKMLSI 315
Query: 309 VVFANQWPLS-----GPTTITLTPSASSSLGPLINAGEVFNVLSLG-GRTSTRDVIALQR 362
+ Q + G + L + S+L PLINA E+F V+ T+ +VIA+++
Sbjct: 316 DTMSPQPDSTLTCKGGSCLLQLVKTTKSTLPPLINAIELFTVVEFPQSETNQDEVIAIKK 375
Query: 363 VKESLRNPPLDWSGDPCVPRQYSWTGITCSE---GLRIRIVTLNLTSMDLSGSLSSFVAN 419
++ + ++W GDPCVP Q+ W G+ CS I LNL+S L+G +S + N
Sbjct: 376 IQLTYGLSRINWQGDPCVPEQFLWAGLKCSNINSSTPPTITFLNLSSSGLTGIISPSIQN 435
Query: 420 MTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSLGNISSLK 471
+T L + L NN L+G +P L+ + L ++L N FSG++P L + LK
Sbjct: 436 LTHLQELDLSNNDLTGDVPEFLADIKSLLIINLSGNNFSGQLPQKLIDKKRLK 488
>At1g51800 unknown protein
Length = 894
Score = 171 bits (434), Expect = 7e-43
Identities = 132/470 (28%), Positives = 241/470 (51%), Gaps = 51/470 (10%)
Query: 25 GFL-INCGTLTTTQIN----NRTWLPDSNFITTGTPKNIT----TQVLLPTLKTLRSFPL 75
GF+ ++CG+ T N T++ D+NFI TG +I TQ T LRSFP
Sbjct: 28 GFISLDCGSPRETSFREKTTNITYISDANFINTGVGGSIKQGYRTQFQQQTWN-LRSFPQ 86
Query: 76 QVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYANG 135
++ +CY + + G +Y+IR + +GG + D P+ F+ + LWS V TT +
Sbjct: 87 GIR-NCYTLNLTIGDEYLIRANFLHGGYD--DKPSTQ-FELYLGPNLWSTVTTTNET--- 139
Query: 136 NSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTD-----FNNFAI 190
+S +E + + + C+ ++ PF+SALE L ++ Y T F +
Sbjct: 140 EASIFEMIHILTTDRLQICLVKTG--NATPFISALELRKLMNTTYLTRQGSLQTFIRADV 197
Query: 191 GLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANT-ENVSVSGFWNLPPSKVFETH 249
G + RY D FDR+W P+ N ++ +T ++V+++ + P +
Sbjct: 198 GATVNQGY-------RYGIDVFDRVWTPYNFGNWSQISTNQSVNINNDYQPPEIAMVTAS 250
Query: 250 LGSEQLESLELRWPTASLPSSKYYIALYFAD----NTAGSRIFNISVNGVHYY---RDLN 302
+ ++ ++ + + ++Y+ ++FA+ + +R FNI N H Y R LN
Sbjct: 251 VPTDPDAAMNISLVGVER-TVQFYVFMHFAEIQELKSNDTREFNIMYNNKHIYGPFRPLN 309
Query: 303 AIASGVV----VFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVLSLGGR-TSTRDV 357
S V V A+ +G +L + +S+L PL+NA E+++V L + T ++V
Sbjct: 310 FTTSSVFTPTEVVADA---NGQYIFSLQRTGNSTLPPLLNAMEIYSVNLLPQQETDRKEV 366
Query: 358 IALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCS--EGLRIRIVTLNLTSMDLSGSLSS 415
A+ +K + +DW GDPCVP Y W+G+ C+ + +I++L+L++ L+G +
Sbjct: 367 DAMMNIKSAYGVNKIDWEGDPCVPLDYKWSGVNCTYVDNETPKIISLDLSTSGLTGEILE 426
Query: 416 FVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSL 464
F++++T+L + L NNSL+G +P L+++ L+ ++L N+ +G IP++L
Sbjct: 427 FISDLTSLEVLDLSNNSLTGSVPEFLANMETLKLINLSGNELNGSIPATL 476
>At5g59680 serine/threonine-specific protein kinase - like
Length = 882
Score = 169 bits (428), Expect = 3e-42
Identities = 136/467 (29%), Positives = 218/467 (46%), Gaps = 54/467 (11%)
Query: 46 DSNFITTGTPKNITTQVLLPTLK---TLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGG 102
D+ FI +G I + LK T+R FP K++CYN+ V +G ++IR + YG
Sbjct: 56 DAAFIQSGKIGRIQANLEADFLKPSTTMRYFP-DGKRNCYNLNVEKGRNHLIRARFVYGN 114
Query: 103 VNGVDHPTPPVFDQIIDGTLWSVVNTTVDYANG-NSSFYEGVFLAVGKFMSFCIGSNSYT 161
+G D T P FD + W+ T+D A N + E + + + C+ T
Sbjct: 115 YDGRD--TGPKFDLYLGPNPWA----TIDLAKQVNGTRPEIMHIPTSNKLQVCLVKTGET 168
Query: 162 DSDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQ 221
P +S LE +G Y T + ++ L R F S S+RYPDD +DR W F
Sbjct: 169 T--PLISVLEVRPMGSGTYLT---KSGSLKLYYREYFSKSDSSLRYPDDIYDRQWTSFFD 223
Query: 222 SNSTKANT-ENVSVSGFWNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFAD 280
+ T+ NT +V S + P + + + L W + + P +YY+ +F++
Sbjct: 224 TEWTQINTTSDVGNSNDYKPPKVALTTAAIPTNASAPLTNEWSSVN-PDEQYYVYAHFSE 282
Query: 281 ----NTAGSRIFNISVNGVHYYRDLN----AIASGVVVFANQWPLSGPTTITLTPSASSS 332
+R FN+ +NG ++ + AI++ + V N G + L + S+
Sbjct: 283 IQELQANETREFNMLLNGKLFFGPVVPPKLAISTILSVSPNTCE-GGECNLQLIRTNRST 341
Query: 333 LGPLINAGEVFNVLSLGG-RTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITC 391
L PL+NA EV+ V+ T+ DV A++ ++ + ++W DPCVP+Q+ W G+ C
Sbjct: 342 LPPLLNAYEVYKVIQFPQLETNETDVSAVKNIQATYELSRINWQSDPCVPQQFMWDGLNC 401
Query: 392 SEG---LRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLET 448
S RI TLNL+S L+G++++ + N LT LE
Sbjct: 402 SITDITTPPRITTLNLSSSGLTGTITAAIQN-----------------------LTTLEK 438
Query: 449 LHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLKPGLSI 495
L L N +GE+P L N+ SL + L N+L G IP +L + GL +
Sbjct: 439 LDLSNNNLTGEVPEFLSNMKSLLVINLSGNDLNGTIPQSLQRKGLEL 485
>At3g46420 hypothetical protein
Length = 838
Score = 168 bits (425), Expect = 7e-42
Identities = 132/435 (30%), Positives = 206/435 (47%), Gaps = 28/435 (6%)
Query: 46 DSNFITTGTPKNITTQVLLPTLK---TLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGG 102
DSNFI TG I + K TLR FP ++ +CYN+ V +G Y+IR YG
Sbjct: 56 DSNFIETGKLGRIQASLEPKYRKSQTTLRYFPDGIR-NCYNLTVTQGTNYLIRARAIYGN 114
Query: 103 VNGVDHPTPPVFDQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTD 162
+G++ P FD I W ++ Y NG ++ E +++ + C+
Sbjct: 115 YDGLN--IYPKFDLYIGPNFWVTIDLG-KYVNG--TWEEIIYIPKSNMLDVCLVKTG--P 167
Query: 163 SDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQS 222
S P +S+L L ++ Y T + + R S IRYPDD +DRIW + +
Sbjct: 168 STPLISSLVLRPLANATYIT---QSGWLKTYVRVYLSDSNDVIRYPDDVYDRIWGSYFEP 224
Query: 223 NSTKANTENVSVSGFWNLPPSKVFETHLGSEQLESLELRWP-TASLPSSKYYIALYFAD- 280
K +T S LPP K T S S L P PS K Y+ L+F++
Sbjct: 225 EWKKISTTLGVNSSSGFLPPLKALMT-AASPANASAPLAIPGVLDFPSDKLYLFLHFSEI 283
Query: 281 ---NTAGSRIFNISVNGVHYYRDLNAIASGVVVFANQWPLS---GPTTITLTPSASSSLG 334
+R F I N Y + + N P++ G + + + S+L
Sbjct: 284 QVLKANETREFEIFWNKKLVYNAYSPVYLQTKTIRNPSPVTCERGECILEMIKTERSTLP 343
Query: 335 PLINAGEVFNVLSLGG-RTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCSE 393
PL+NA EVF V+ T DV+A++ +K + W GDPCVP+Q+ W G+ C+
Sbjct: 344 PLLNAVEVFTVVEFPQPETDASDVVAIKNIKAIYGLTRVTWQGDPCVPQQFLWNGLNCNS 403
Query: 394 ---GLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETL 449
RI +L+L+S L+GS+S + N+T L + L NN+L+G++P+ L+++ L +
Sbjct: 404 METSTPPRITSLDLSSSGLTGSISVVIQNLTHLEKLDLSNNNLTGEVPDFLANMKFLVFI 463
Query: 450 HLEENQFSGEIPSSL 464
+L +N +G IP +L
Sbjct: 464 NLSKNNLNGSIPKAL 478
>At1g51890 Putative protein kinase
Length = 876
Score = 166 bits (420), Expect = 3e-41
Identities = 144/496 (29%), Positives = 241/496 (48%), Gaps = 39/496 (7%)
Query: 25 GFL-INCGTLTTT-----QINNRTWLPDSNFITTGTPKNIT----TQVLLPTLKTLRSFP 74
GF+ ++CG + T + N T+ D+ +I +G P I TQ + LRSFP
Sbjct: 24 GFISLDCGLVPTEITYVEKSTNITYRSDATYIDSGVPGKINEVYRTQ-FQQQIWALRSFP 82
Query: 75 LQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYAN 134
+ +++CYN + KY+IR T+ YG +G++ P FD I W+ V+
Sbjct: 83 -EGQRNCYNFSLTAKRKYLIRGTFIYGNYDGLNQL--PSFDLYIGPNKWTSVSIP---GV 136
Query: 135 GNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVA 194
N S E + + + C+ T PF+S+LE L ++ Y T + ++ +VA
Sbjct: 137 RNGSVSEMIHVLRQDHLQICLVKTGETT--PFISSLELRPLNNNTYVT---KSGSLIVVA 191
Query: 195 RNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTE-NVSVSGFWNLPPSKVFETHLGSE 253
R F + P +RY +D DRIW PF + ++ +TE +V S F+N+P + +
Sbjct: 192 RLYFSPTPPFLRYDEDVHDRIWIPFLDNKNSLLSTELSVDTSNFYNVPQTVAKTAAVPLN 251
Query: 254 QLESLELRWPTASLPSSKYYIALYFAD----NTAGSRIFNISVNG----VHYYRDLNAIA 305
+ L++ W + +S+ YI ++FA+ +R FNI+ NG Y+R
Sbjct: 252 ATQPLKINWSLDDI-TSQSYIYMHFAEIENLEANETREFNITYNGGENWFSYFRPPKFRI 310
Query: 306 SGVVVFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVLSLGGRTSTRD-VIALQRVK 364
+ V A L G T + + +S+ PLIN E++ VL L + +D V A+ +K
Sbjct: 311 TTVYNPAAVSSLDGNFNFTFSMTGNSTHPPLINGLEIYQVLELPQLDTYQDEVSAMMNIK 370
Query: 365 ESLR-NPPLDWSGDPCVPRQYSWTGITCS--EGLRIRIVTLNLTSMDLSGSLSSFVANMT 421
+ W GDPC P Y W G+ CS +I++LNL+ +LSG+++S ++ +T
Sbjct: 371 TIYGLSKRSSWQGDPCAPELYRWEGLNCSYPNFAPPQIISLNLSGSNLSGTITSDISKLT 430
Query: 422 ALTNIWLGNNSLSGQIPNL-SSLTMLETLHLEENQ-FSGEIPSSLGNISSLKEVFLQNNN 479
L + L NN LSG IP + S + L ++L N+ + +P +L K + L +
Sbjct: 431 HLRELDLSNNDLSGDIPFVFSDMKNLTLINLSGNKNLNRSVPETLQKRIDNKSLTLIRDE 490
Query: 480 LTGQIPANLLKPGLSI 495
TG+ N++ S+
Sbjct: 491 -TGKNSTNVVAIAASV 505
>At3g46350 receptor-like protein kinase homolog
Length = 871
Score = 165 bits (417), Expect = 6e-41
Identities = 137/462 (29%), Positives = 230/462 (49%), Gaps = 53/462 (11%)
Query: 24 KGFL-INCGTLTTTQ------INNRTWLPDSNFITTGTPKNITTQVLLPTLK---TLRSF 73
+GF+ ++CG T + + DSNFI +G I T + LK TLR F
Sbjct: 28 EGFISLDCGLAPTEPSPYTEPVTTLQYSSDSNFIQSGKLGRIDTSLQTFFLKQQTTLRYF 87
Query: 74 PLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYA 133
P ++ +CYN+ V +G Y+IR + YG +G + P FD + LW ++ T
Sbjct: 88 PDGIR-NCYNLTVKQGTNYLIRARFTYGNYDGRN--MSPTFDLYLGPNLWKRIDMT-KLQ 143
Query: 134 NGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLV 193
N S+ E ++ + + C+ + T PF+SALE L + Y TT + +
Sbjct: 144 NKVSTLEEITYIPLSNSLDVCLVKTNTTI--PFISALELRPLPSNSYITTAGS---LRTF 198
Query: 194 ARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTE-NVSVSGFWNLPPSKVFETHLGS 252
R F S IR+P D DR+WE + + T+ +T V+ S + LP + + +
Sbjct: 199 VRFCFSNSVEDIRFPMDVHDRMWESYFDDDWTQISTSLTVNTSDSFRLPQAALITAATPA 258
Query: 253 EQLES-LELRWPTASLPSSKYYIALYFADNTA----GSRIFNISVNGVHYYRDLNAIASG 307
+ S + + + T+S +++I L+F++ A +R FNIS+NG ++A
Sbjct: 259 KDGPSYIGITFSTSS--EERFFIYLHFSEVQALRANETREFNISING-------ESVAD- 308
Query: 308 VVVFANQWPLSGPTTITLTPSASSSLGPLINAGEVFNVLSL-GGRTSTRDVIALQRVKES 366
L P L+ + SS+ P+INA E+F V L T DVIA++++K++
Sbjct: 309 ---------LYRP----LSRTQSSTHPPMINAIEIFLVSELLQSETYENDVIAIKKIKDT 355
Query: 367 LRNPPLDWSGDPCVPRQYSWTGITCSEG---LRIRIVTLNLTSMDLSGSLSSFVANMTAL 423
+ W GDPCVPR Y W G+ C++ + RI +L L+S L+G++++ + +T+L
Sbjct: 356 YGLQLISWQGDPCVPRLYKWDGLDCTDTDTYIAPRITSLKLSSKGLTGTIAADIQYLTSL 415
Query: 424 TNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSGEIPSSL 464
+ L +N L G +P L+++ L ++L +N G IP +L
Sbjct: 416 EKLDLSDNKLVGVVPEFLANMKSLMFINLTKNDLHGSIPQAL 457
>At1g51810 Putative protein kinase
Length = 871
Score = 164 bits (415), Expect = 1e-40
Identities = 136/493 (27%), Positives = 233/493 (46%), Gaps = 44/493 (8%)
Query: 4 LFFLLLFISFHTPSFSQTPPKGFL-INCGTLTT-----TQINNRTWLPDSNFITTGTPKN 57
LFF++ + H P GF+ ++CG T+ D F+ +G
Sbjct: 6 LFFVIFSLILHLVQAQD--PIGFINLDCGLSIQGSPYKESSTGLTYTSDDGFVQSGKIGK 63
Query: 58 ITTQV---LLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVF 114
IT ++ +TLR FP V+ +C+++ V RG KY+I+ T+ YG +G + P F
Sbjct: 64 ITKELESLYKKPERTLRYFPDGVR-NCFSLNVTRGTKYLIKPTFLYGNYDGRN--VIPDF 120
Query: 115 DQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLI 174
D I +W VNT +++ E + ++ + C+ S P+++ LE
Sbjct: 121 DLYIGPNMWITVNT-------DNTIKEILHVSKSNTLQVCLVKTG--TSIPYINTLELRP 171
Query: 175 LGDSLY-NTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTEN-- 231
L D +Y N + N+ + N GY I YPDD DRIW+ + T N
Sbjct: 172 LADDIYTNESGSLNYLFRVYYSNLKGY----IEYPDDVHDRIWKQILPYQDWQILTTNLQ 227
Query: 232 VSVSGFWNLPPSKVFETHLGSEQLESLELRWP-TASLPSSKYYIALYFAD----NTAGSR 286
++VS ++LP +V +T + + + + +P P+S++Y+ L+FA+ +R
Sbjct: 228 INVSNDYDLP-QRVMKTAVTPIKASTTTMEFPWNLEPPTSQFYLFLHFAELQSLQANETR 286
Query: 287 IFNISVNGVHYYRDLNAIASGVVVFANQWPLS---GPTTITLTPSASSSLGPLINAGEVF 343
FN+ +NG ++ + + + P G + L ++ S+L PLINA E +
Sbjct: 287 EFNVVLNGNVTFKSYSPKFLEMQTVYSTAPKQCDGGKCLLQLVKTSRSTLPPLINAMEAY 346
Query: 344 NVLSLGG-RTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTGITCS---EGLRIRI 399
VL T+ +VIA++ ++ + W GDPCVP+++ W G+ C+ + I
Sbjct: 347 TVLDFPQIETNVDEVIAIKNIQSTYGLSKTTWQGDPCVPKKFLWDGLNCNNSDDSTPPII 406
Query: 400 VTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEENQFSG 458
+LNL+S L+G + + N+ L + L NN+LSG +P L+ + L ++L N SG
Sbjct: 407 TSLNLSSSGLTGIIVLTIQNLANLQELDLSNNNLSGGVPEFLADMKSLLVINLSGNNLSG 466
Query: 459 EIPSSLGNISSLK 471
+P L LK
Sbjct: 467 VVPQKLIEKKMLK 479
>At1g51910 Putative protein kinase
Length = 879
Score = 163 bits (413), Expect = 2e-40
Identities = 143/509 (28%), Positives = 251/509 (49%), Gaps = 51/509 (10%)
Query: 5 FFLLLFISFHTPSFSQTPPK-GFL-INCG-----TLTTTQINNRTWLPDSNFITTGTPKN 57
F LL I+F Q + GF+ ++CG T T QI N T++ D+++I +G +
Sbjct: 7 FLLLSTIAFAVFHLVQAQSQSGFISLDCGLIPKDTTYTEQITNITYISDADYIDSGLTER 66
Query: 58 ITTQV---LLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVF 114
I+ L TLRSFP + +++CYN + KY+IR T+ YG +G++ P F
Sbjct: 67 ISDSYKSQLQQQTWTLRSFP-EGQRNCYNFNLKANLKYLIRGTFVYGNYDGLNQM--PKF 123
Query: 115 DQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGSNSYTDSDPFVSALEFLI 174
D I W+ V + N++ +E + + + C+ T PF+S+LE
Sbjct: 124 DLHIGPNKWTSV---ILEGVANATIFEIIHVLTQDRLQVCLVKTGQTT--PFISSLELRP 178
Query: 175 LGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWEPFGQSNSTKANTENVSV 234
L + Y T + + AR F + +RY DD +DR+W PF Q+ + +T N+ V
Sbjct: 179 LNNDTYVTQGGSLMSF---ARIYFPKTAYFLRYSDDLYDRVWVPFSQNETVSLST-NLPV 234
Query: 235 ---SGFWNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIALYFAD----NTAGSRI 287
S +N+P + + +E L + W ++ ++ Y+ ++FA+ R
Sbjct: 235 DTSSNSYNVPQNVANSAIIPAEATHPLNIWWDLQNI-NAPSYVYMHFAEIQNLKANDIRE 293
Query: 288 FNISVNGVHYYRD--------LNAIASGVVVFANQWPLSGPTTITLTPSASSSLGPLINA 339
FNI+ NG + + I+S + ++ G T T + +S+L PLINA
Sbjct: 294 FNITYNGGQVWESSIRPHNLSITTISSPTALNSSD----GFFNFTFTMTTTSTLPPLINA 349
Query: 340 GEVFNVL-SLGGRTSTRDVIALQRVKESLR-NPPLDWSGDPCVPRQYSWTGITC--SEGL 395
EV+ ++ +L T +V A+ +K++ + + W GDPC P+ Y W G+ C +
Sbjct: 350 LEVYTLVENLLLETYQDEVSAMMNIKKTYGLSKKISWQGDPCSPQIYRWEGLNCLYLDSD 409
Query: 396 RIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLETLHLEEN 454
+ I +LNL + L+G ++ ++N+ L + L +N LSG+IP+ L+ + ML ++L+ N
Sbjct: 410 QPLITSLNLRTSGLTGIITHDISNLIQLRELDLSDNDLSGEIPDFLADMKMLTLVNLKGN 469
Query: 455 -QFSGEIPSSLG---NISSLKEVFLQNNN 479
+ + +P S+ N SLK + +N +
Sbjct: 470 PKLNLTVPDSIKHRINNKSLKLIIDENQS 498
>At1g25570 hypothetical protein
Length = 628
Score = 163 bits (413), Expect = 2e-40
Identities = 155/523 (29%), Positives = 250/523 (47%), Gaps = 53/523 (10%)
Query: 5 FFLLLFISFHTPSFSQTPPKGFLINCGTL-TTTQINNRTWLPDSNFITTGTPKNITTQVL 63
FFLLL +S +P + F I+CG+ T+T + NRTWLPD F + G+ ++ +
Sbjct: 14 FFLLLHLSLSSPFNTS-----FFIDCGSPETSTDVFNRTWLPDQ-FYSGGSTAVVSEPLR 67
Query: 64 LPTL--KTLRSFPLQV-KKHCYNIPVYRGAKYMIRTTYFYGGVNGVDHPTPPVFDQIIDG 120
+ KT+R FPL KK+CY +P+ G +Y +RT Y +G H P FD ++G
Sbjct: 68 FHLIAEKTIRYFPLSFGKKNCYVVPLPPG-RYYLRTFTVYDNYDGKSHS--PSFDVSVEG 124
Query: 121 TLWSVVNTTVDYANGNSSFYEGVFLAVGKF-MSFCIGSNSYTDSDPFVSALEFLILGDSL 179
TL + + Y +F +G + C S P V +LE L + S
Sbjct: 125 TLVFSWRSPWPESLLRDGSYSDLFAFIGDGELDLCF--YSIATDPPIVGSLEVLQVDPSS 182
Query: 180 YNTTDFN-NFAIGLVARNSFGYS--GPSIRYPDDQFDRIW---EPFGQSNSTKANTENVS 233
Y+ N + R S G GP D F R W E F +S +++ ++S
Sbjct: 183 YDADGTGQNVLLVNYGRLSCGSDQWGPGFTNHTDNFGRSWQSDEDF-RSEDSRSVARSLS 241
Query: 234 ----VSGFWNLP---PSKVFETHL----GSEQLESLELRWPTASLPSSKYYIALYFAD-- 280
+ G P P K+++T + G + LE+ A L Y + +F++
Sbjct: 242 TLEKIKGVDQAPNYFPMKLYQTAVTVSGGGSLVYELEV---DAKLD---YLLWFHFSEID 295
Query: 281 ---NTAGSRIFNISVNGVHYYR-DLNAIASGVVVFANQWPLSGPTTITLTPSASSSLG-P 335
AG R+F++ VN + R D+ G ++ + + ++ +T SS G P
Sbjct: 296 STVKKAGQRVFDLVVNDNNVSRVDVFHEVGGFAAYSLNYTVKNLSSTIVTVKLSSVSGAP 355
Query: 336 LINAGEVFNVLSLGGRTSTRDVIALQRVKESLRNPP-LDWSGDPCVPRQY-SWTGITC-- 391
+I+ E + ++ T V A++ +K+SLR P + W+GDPC P + +W G++C
Sbjct: 356 IISGLENYAIVPADMATVPEQVTAMKALKDSLRVPDRMGWNGDPCAPTSWDAWEGVSCRP 415
Query: 392 -SEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPNLSSLTMLETLH 450
S+G + I ++L S L G +S ++ +T L ++ L +N+LSGQ+P L +L
Sbjct: 416 NSQGSALVIFQIDLGSQGLKGFISEQISLLTNLNSLNLSSNTLSGQLPLGLGHKSLVSLD 475
Query: 451 LEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLLKPGL 493
L NQ +G IP SL +SSLK V L N L G++P + G+
Sbjct: 476 LSNNQLTGPIPESL-TLSSLKLVLLNGNELQGKVPEEVYSVGV 517
>At1g51820 Putative protein kinase
Length = 883
Score = 161 bits (408), Expect = 7e-40
Identities = 124/469 (26%), Positives = 225/469 (47%), Gaps = 53/469 (11%)
Query: 42 TWLPDSNFITTGTPKNITTQ----VLLPTLKTLRSFPLQVKKHCYNIPVYRGAKYMIRTT 97
T+ D++ + +G + + V PTL TLR FP V+ +CYN+ V Y+I+ T
Sbjct: 48 TYTSDADLVASGKTGRLAKEFEPLVDKPTL-TLRYFPEGVR-NCYNLNVTSDTNYLIKAT 105
Query: 98 YFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYANGNSSFYEGVFLAVGKFMSFCIGS 157
+ YG +G++ P F+ + LW+ V++ N + E + + + C+
Sbjct: 106 FVYGNYDGLN--VGPNFNLYLGPNLWTTVSS-------NDTIEEIILVTRSNSLQVCLVK 156
Query: 158 NSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDRIWE 217
S PF++ LE + ++Y T + ++ + R S IR+PDD +DR W
Sbjct: 157 TGI--SIPFINMLELRPMKKNMYVT---QSGSLKYLFRGYISNSSTRIRFPDDVYDRKWY 211
Query: 218 PFGQSNSTKANTE-NVSVSGFWNLPPSKVFETHLGSEQLESLELRWPTASLPSSKYYIAL 276
P + T+ T V+ S + LP S + + + ++L + W T P++++Y +
Sbjct: 212 PLFDDSWTQVTTNLKVNTSITYELPQSVMAKAATPIKANDTLNITW-TVEPPTTQFYSYV 270
Query: 277 YFADNTA----GSRIFNISVNGVHYYRDLNAI---ASGVVVFANQWPLSGPTTITLTPSA 329
+ A+ A +R FN+++NG + + + I + +V + G + + +
Sbjct: 271 HIAEIQALRANETREFNVTLNGEYTFGPFSPIPLKTASIVDLSPGQCDGGRCILQVVKTL 330
Query: 330 SSSLGPLINAGEVFNVLSLGG-RTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQYSWTG 388
S+L PL+NA E F V+ T+ DV ++ V+ + + W GDPCVP+Q W G
Sbjct: 331 KSTLPPLLNAIEAFTVIDFPQMETNENDVAGIKNVQGTYGLSRISWQGDPCVPKQLLWDG 390
Query: 389 ITCSEGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN-LSSLTMLE 447
+ C +S ++ +T++ L ++ L+G I + +LT L+
Sbjct: 391 LNCK---------------------NSDISTPPIITSLDLSSSGLTGIITQAIKNLTHLQ 429
Query: 448 TLHLEENQFSGEIPSSLGNISSLKEVFLQNNNLTGQIPANLL-KPGLSI 495
L L +N +GE+P L +I SL + L NNL+G +P +LL K G+++
Sbjct: 430 ILDLSDNNLTGEVPEFLADIKSLLVINLSGNNLSGSVPPSLLQKKGMNV 478
>At1g07550 protein kinase like
Length = 864
Score = 161 bits (408), Expect = 7e-40
Identities = 127/445 (28%), Positives = 211/445 (46%), Gaps = 33/445 (7%)
Query: 39 NNRTWLPDSNFITTGTPKNITTQVLLPTLK---TLRSFPLQVKKHCYNIPVYRGAKYMIR 95
+N T++ D++FI G N+ +L+ K LR FP ++ +CY++ V + Y+IR
Sbjct: 48 SNLTYISDADFIQGGKTGNVQKDLLMKLRKPYTVLRYFPDGIR-NCYSLNVKQDTNYLIR 106
Query: 96 TTYFYGGVNGVDHPTPPVFDQIIDGTLWSVVNTTVDYA-NGNSSFYEGVFLAVGKFMSFC 154
+ YG +G+++ P FD + +W TT+D +G+ E + + + C
Sbjct: 107 VMFRYGNYDGLNNS--PRFDLYLGPNIW----TTIDMGKSGDGVLEEIIHITRSNILDIC 160
Query: 155 IGSNSYTDSDPFVSALEFLILGDSLYNTTDFNNFAIGLVARNSFGYSGPSIRYPDDQFDR 214
+ S P +S++E L LY+T ++ R F S IRYP D DR
Sbjct: 161 LVKTG--TSTPMISSIELRPL---LYDTYIAQTGSLRNYNRFYFTDSNNYIRYPQDVHDR 215
Query: 215 IWEPFGQSNSTKANTENV---SVSGFWNLPPSKVFETH-LGSEQLESLELRWPTASLPSS 270
IW P T NT + S+ G+ PP V T + + + + + W +
Sbjct: 216 IWVPLILPEWTHINTSHHVIDSIDGYD--PPQDVLRTGAMPANASDPMTITWNLKTATDQ 273
Query: 271 KY---YIALYFADNTAGSRIFNISVNGVHYYRDLNAIASGVVVFANQWPLS---GPTTIT 324
Y YIA +R F + VN ++ V N PL+ G +
Sbjct: 274 VYGYIYIAEIMEVQANETREFEVVVNNKVHFDPFRPTRFEAQVMFNNVPLTCEGGFCRLQ 333
Query: 325 LTPSASSSLGPLINAGEVFNVLSLG-GRTSTRDVIALQRVKESLRNPPLDWSGDPCVPRQ 383
L + S+L PL+NA E+F + T+ DVIA++ ++ S + W GDPCVP+Q
Sbjct: 334 LIKTPKSTLPPLMNAFEIFTGIEFPQSETNQNDVIAVKNIQASYGLNRISWQGDPCVPKQ 393
Query: 384 YSWTGITCS---EGLRIRIVTLNLTSMDLSGSLSSFVANMTALTNIWLGNNSLSGQIPN- 439
+ WTG++C+ RIV L+L+S L+G + + N+T L + L N+L+G++P
Sbjct: 394 FLWTGLSCNVIDVSTPPRIVKLDLSSSGLNGVIPPSIQNLTQLQELDLSQNNLTGKVPEF 453
Query: 440 LSSLTMLETLHLEENQFSGEIPSSL 464
L+ + L ++L N+ SG +P +L
Sbjct: 454 LAKMKYLLVINLSGNKLSGLVPQAL 478
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,613,296
Number of Sequences: 26719
Number of extensions: 517464
Number of successful extensions: 6731
Number of sequences better than 10.0: 434
Number of HSP's better than 10.0 without gapping: 393
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 1180
Number of HSP's gapped (non-prelim): 2767
length of query: 496
length of database: 11,318,596
effective HSP length: 103
effective length of query: 393
effective length of database: 8,566,539
effective search space: 3366649827
effective search space used: 3366649827
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC137838.4


